-

Figure 1.
Fruit shapes and seed sizes of the seeded and seedless Diospyros lotus. (a) Seeded (Yz01); (b) Seedless (W01).
-
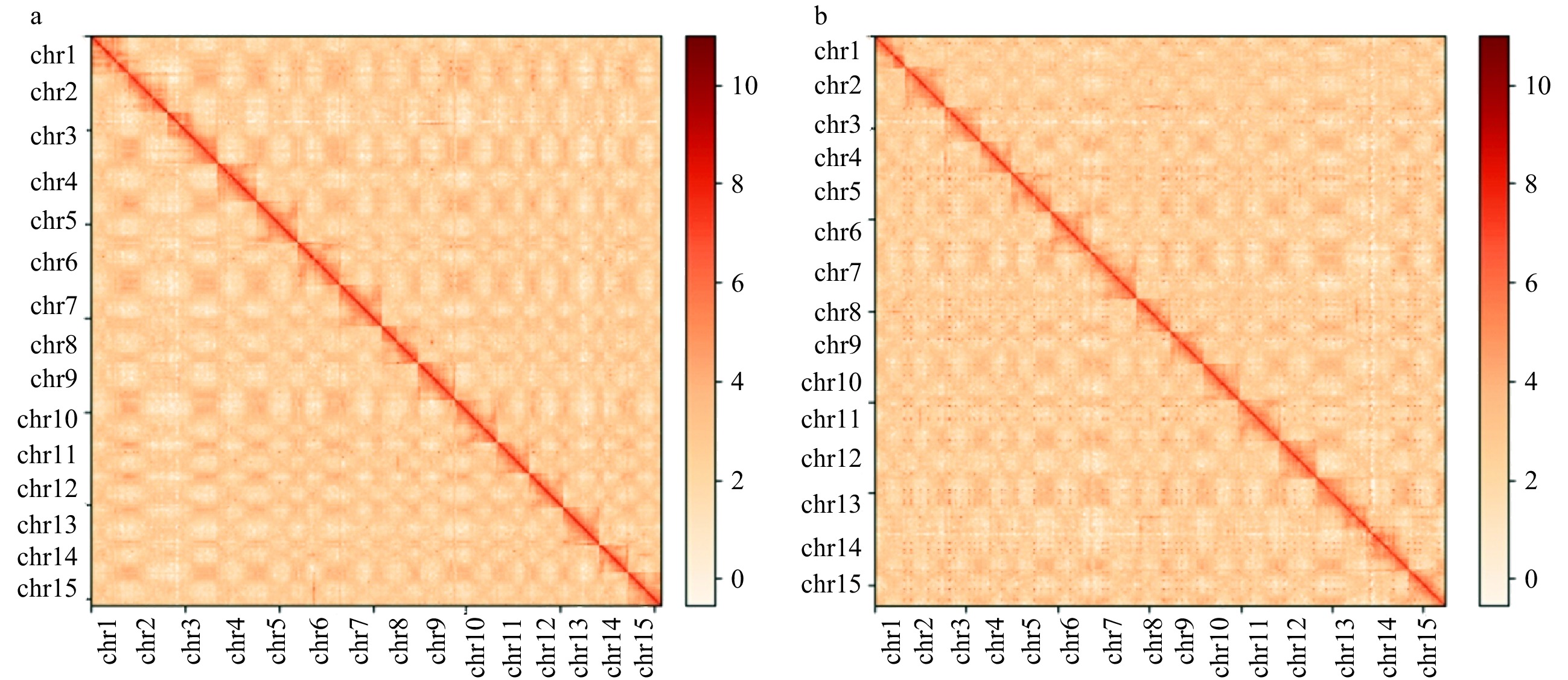
Figure 2.
Hi-C interaction heat maps for Diospyros lotus genomes presenting the interactions among 15 chromosomes. (a) Seeded (Yz01); (b) Seedless (W01).
-
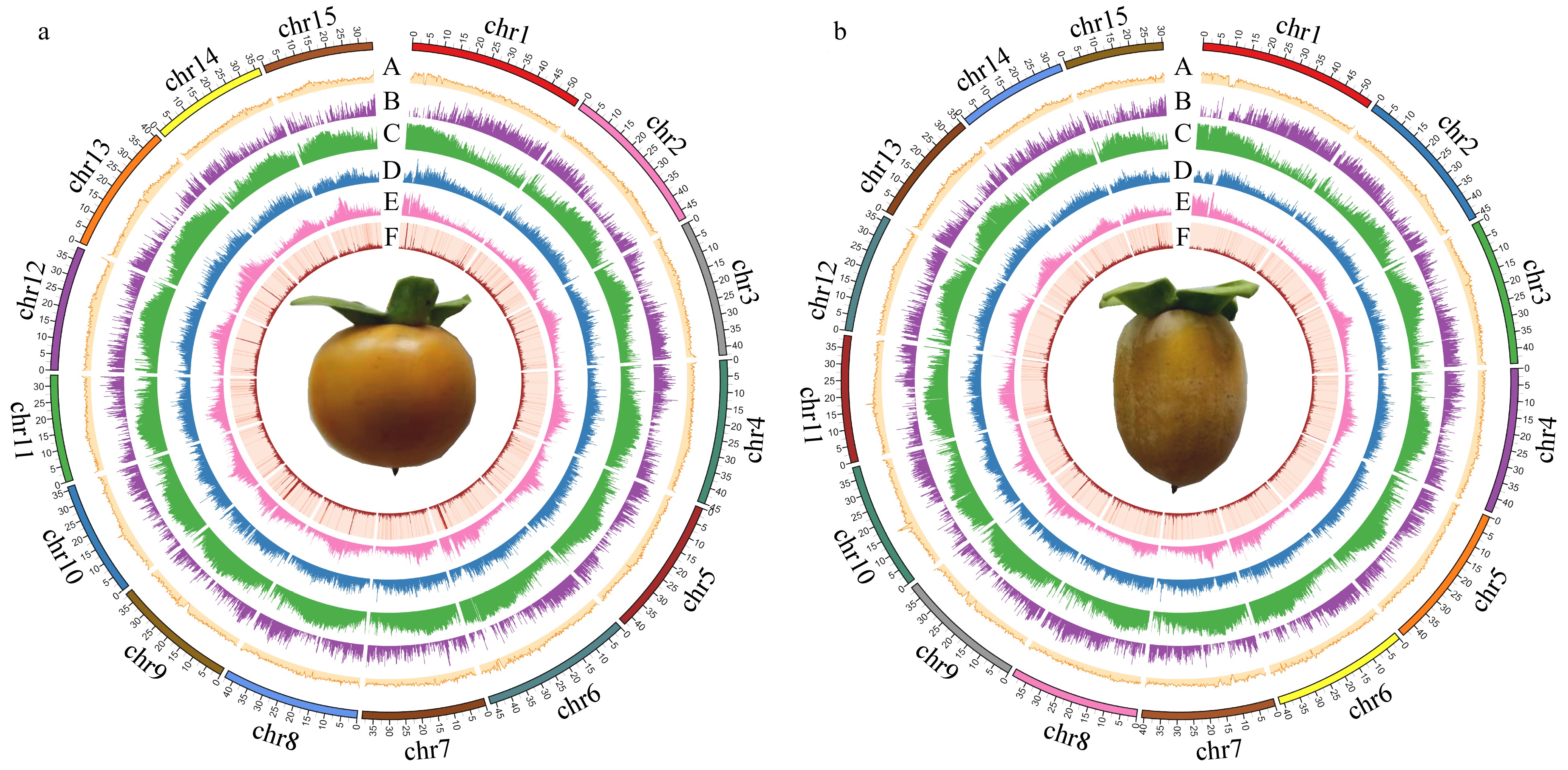
Figure 3.
Circular genomic maps for Diospyros lotus. (a) Seeded (Yz01); (b) Seedless (W01). A. GC content distribution; B. Gene density distribution; C. Repeats density distribution; D. LTR-Copia density distribution; E. LTR-Gypsy density distribution; F. DNA transposon density distribution.
-
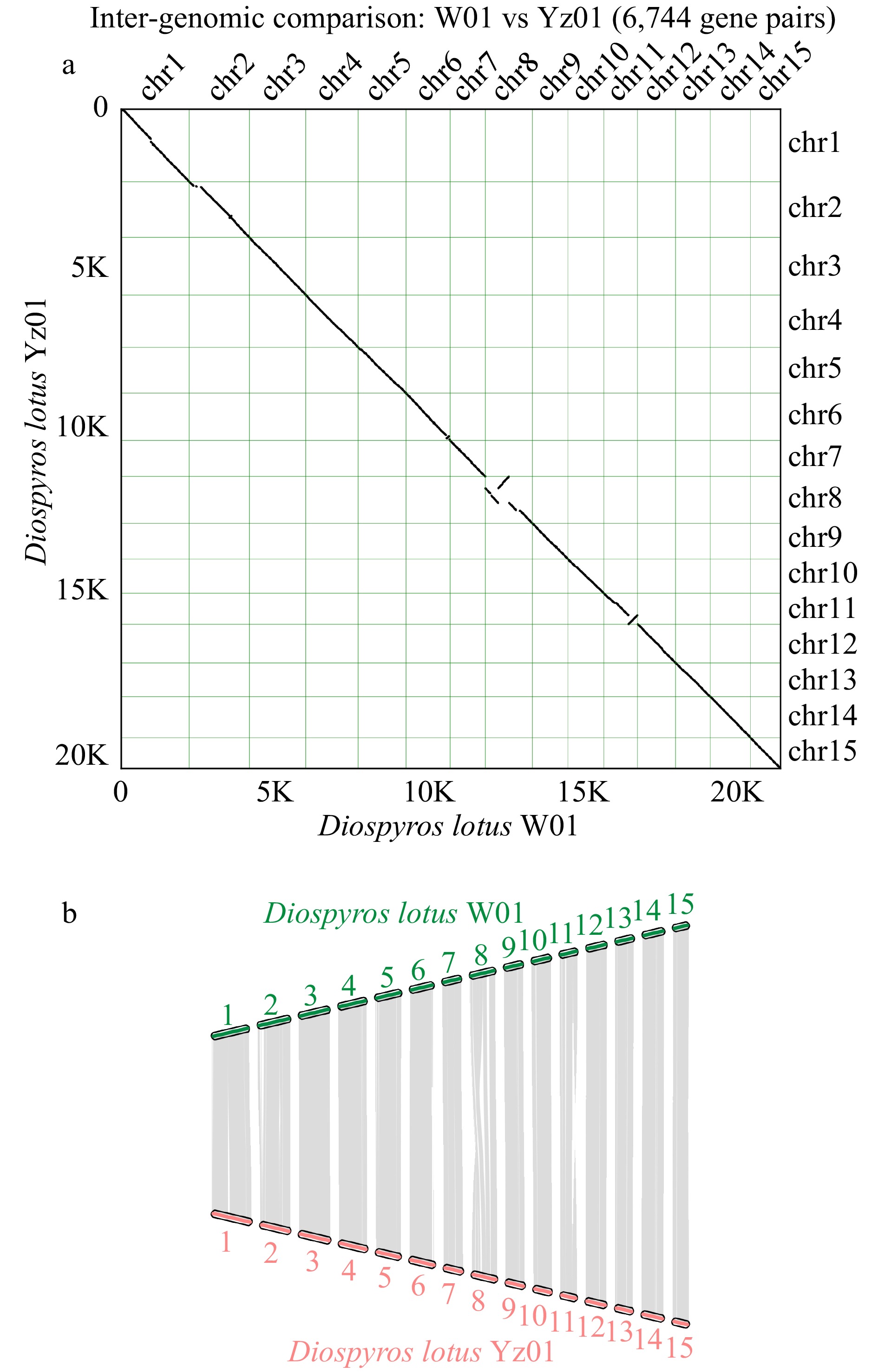
Figure 4.
Chromosomal synteny between seedless and seeded Diospyros lotus. (a) Inter-genomic comparison. (b) Chromosomal maps of seedless and seeded Diospyros lotus.
-
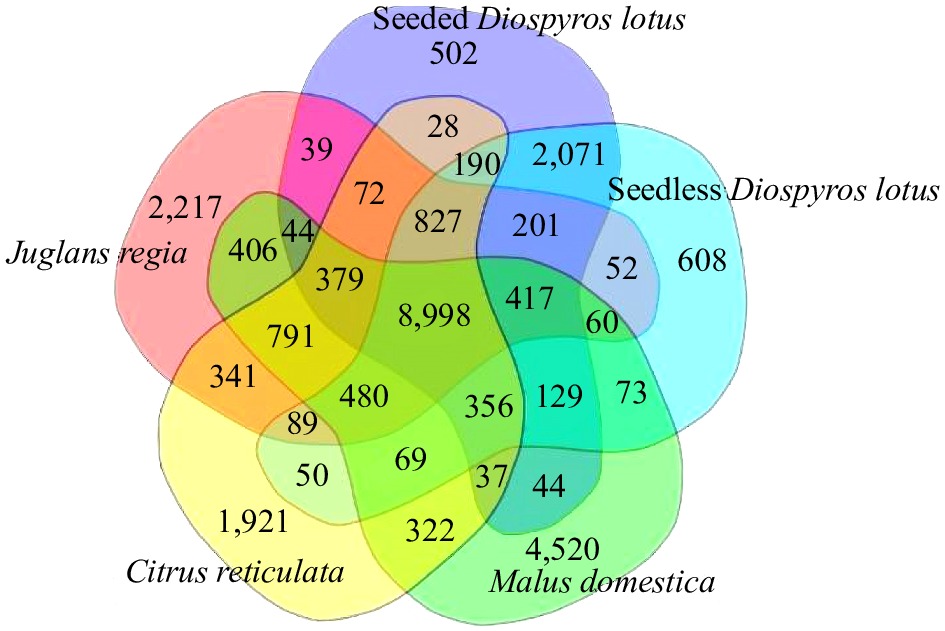
Figure 5.
Venn diagram presenting the number of shared and unique protein-coding genes among seedless and seeded Diospyros lotus, Malus domestica, Citrus reticulata, and Juglans regia revealed by an orthology analysis.
-
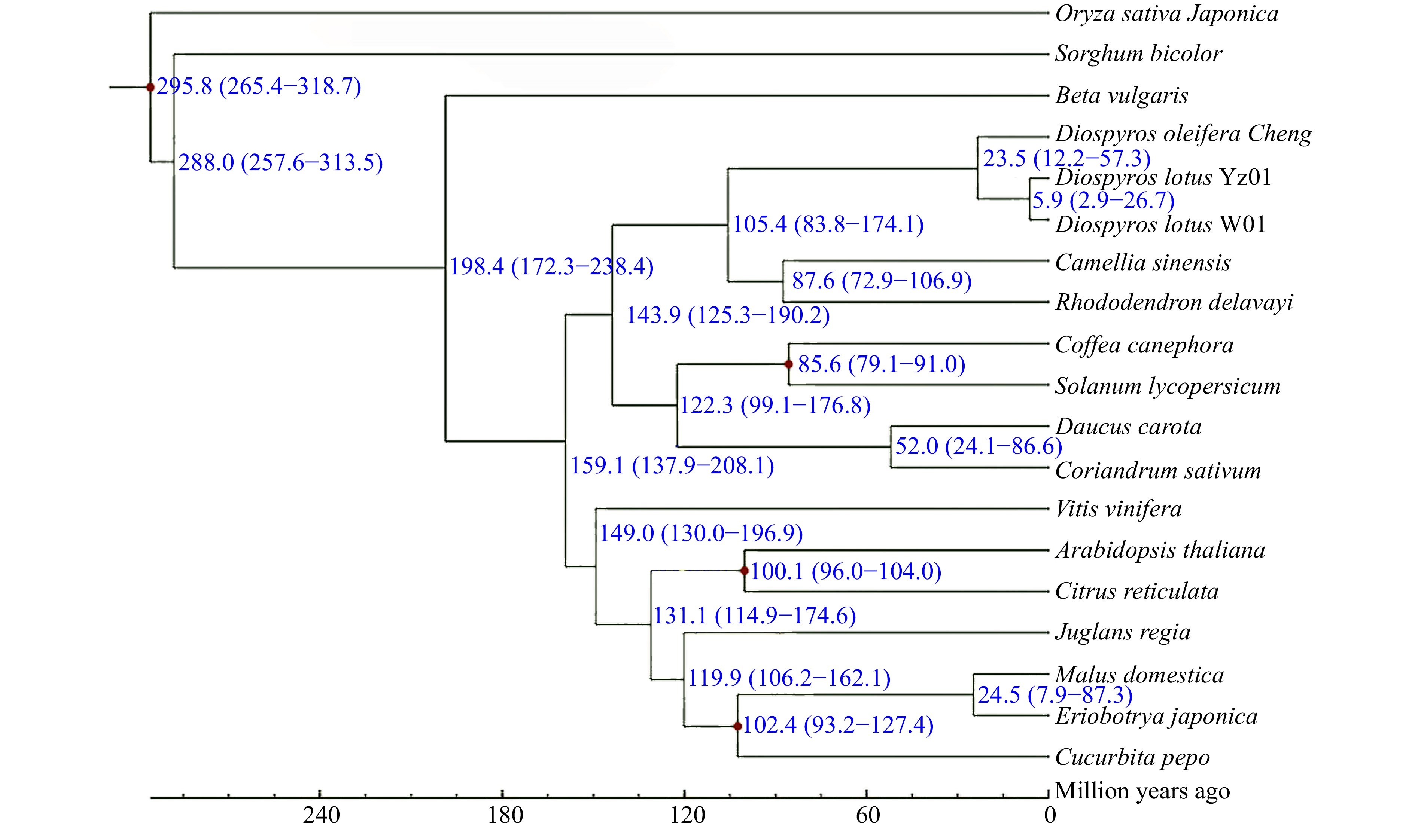
Figure 6.
Phylogenetic tree of seedless and seeded Diospyros lotus and 17 other species constructed using the maximum-likelihood method. Estimated species divergence times (million years ago) and 95% confidence intervals are labeled at each branch site. Blue numbers on branches indicate the estimated divergence times. Red dots indicate the divergence times estimated based on fossil evidence.
-
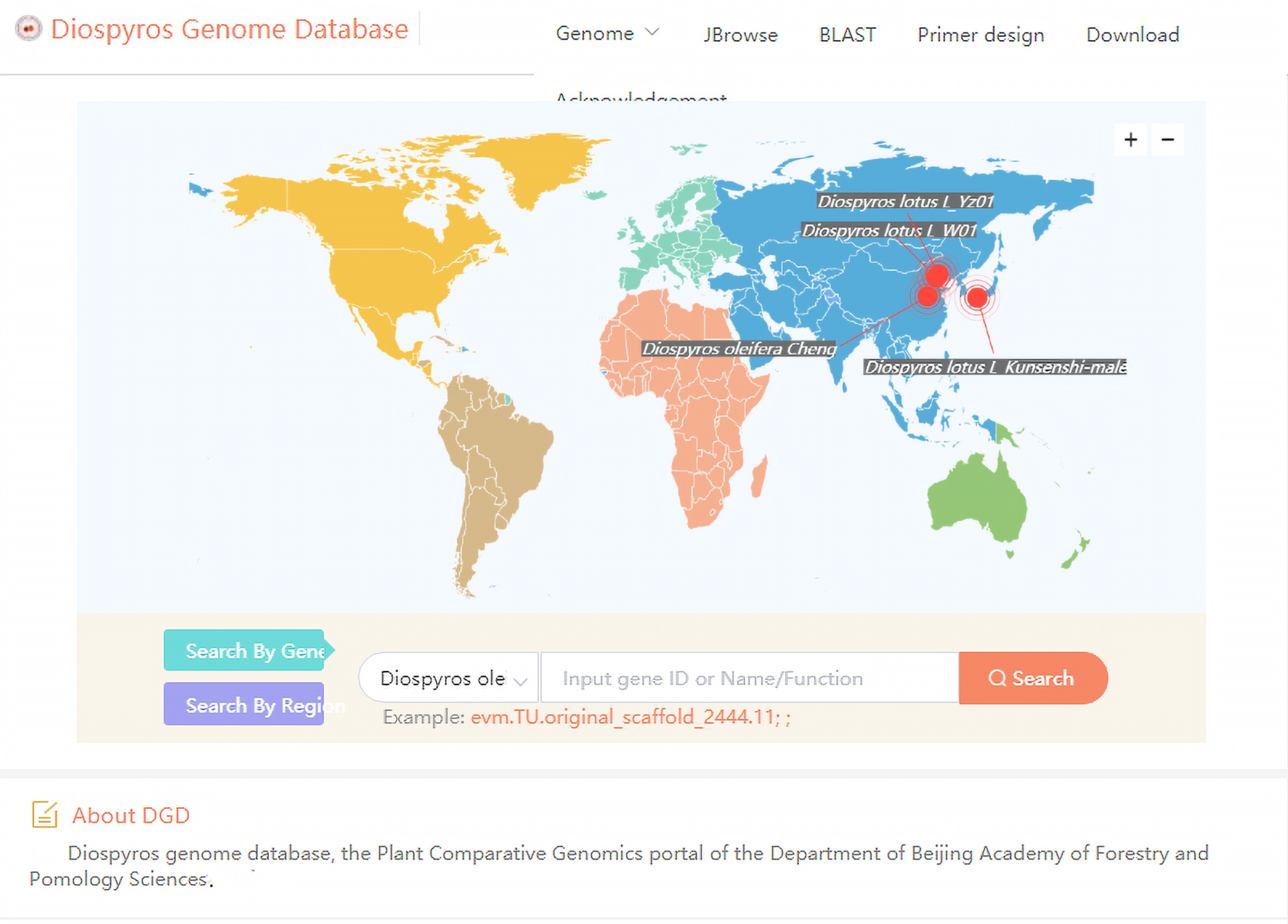
Figure 7.
The user interface of Diospyros Genome Database for browsing genomes, searching for homologous sequences and designing primers.
-
Library type Seedless Diospyros lotus (W01) Seeded Diospyros lotus (Yz01) Library size (bp) Clean data (Gb) Coverage (×) Library size (bp) Clean data (Gb) Coverage (×) Illumina 350 80.99 119.53 350 79.21 114.98 Pacbio 20,000 92.1 103.29 20,000 133.51 166.98 Hi-C 350 86.96 − 350 107.87 − Table 1.
Summary of the sequencing data used for assembling the Diospyros lotus genomes.
-
Parameter Seedless Diospyros lotus (W01) Seeded Diospyros lotus (Yz01) Contig length (bp) Contig number Contig length (bp) Contig number N90 561,232 228 537,928 279 N80 1,144,354 151 1,078,450 194 N70 1,625,012 106 1,450,541 143 N60 2,258,638 73 2,059,392 106 N50 3,006,748 49 2,463,960 77 Total length 617,662,490 − 647,313,630 − Number (≥ 100 bp) − 706 − 743 Number (≥ 2 kb) − 691 − 734 Max length 16,262,241 − 14,842,567 − Table 2.
Summary of the assembled seedless and seeded Diospyros lotus genomes.
-
Type Seedless Diospyros lotus (W01) Seeded Diospyros lotus (Yz01) Number Percent (%) Number Percent (%) Total 21,684 − 23,193 − Annotated 20,689 95.41 22,844 98.5 InterPro 17,473 80.58 20,037 86.39 GO 12,161 56.08 14,066 60.65 KEGG ALL 20,547 94.76 22,750 98.09 KEGG KO 8,435 38.90 9,812 42.31 Swissprot 15,057 69.44 16,896 72.85 TrEMBL 20,587 94.94 22,790 98.26 TF 1,560 7.19 1,572 6.78 Pfam 17,064 78.69 19,709 84.98 NR 20,607 95.03 22,794 98.28 KOG 17,990 82.96 20,208 87.13 Unannotated − 995 4.59 349 1.50 Table 3.
General statistics for the functional annotations of the genes in the seedless and seeded Diospyros lotus genomes.
Figures
(7)
Tables
(3)