-
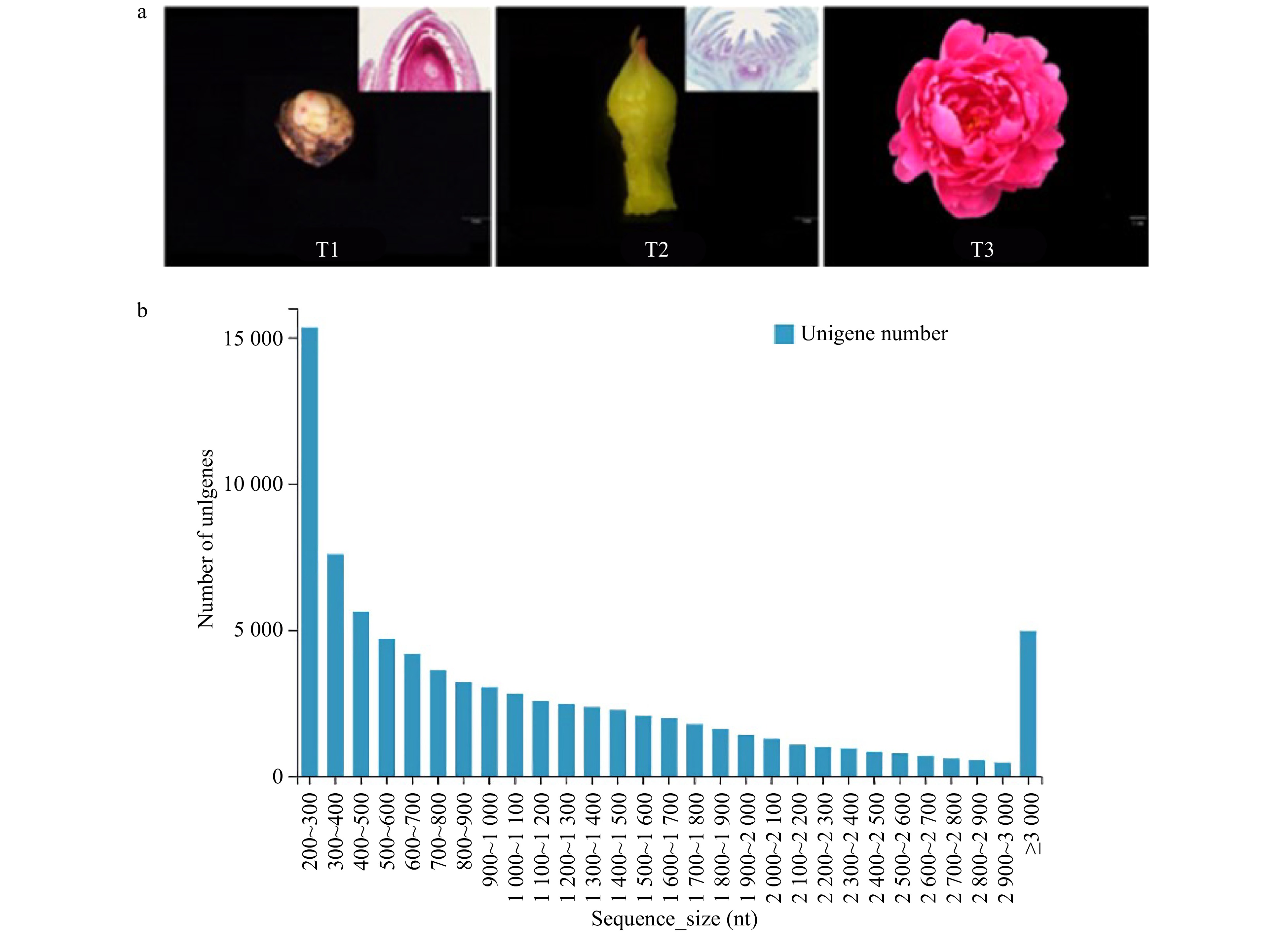
Figure 1. Transcriptome sequencing of 'Dafugui'. (a) Materials of 'Dafugui' in three periods. T1, critical period of flower bud differentiation; T2, flower bud morphogenesis; T3, peak flowering. (b) Unigene length distribution.
-
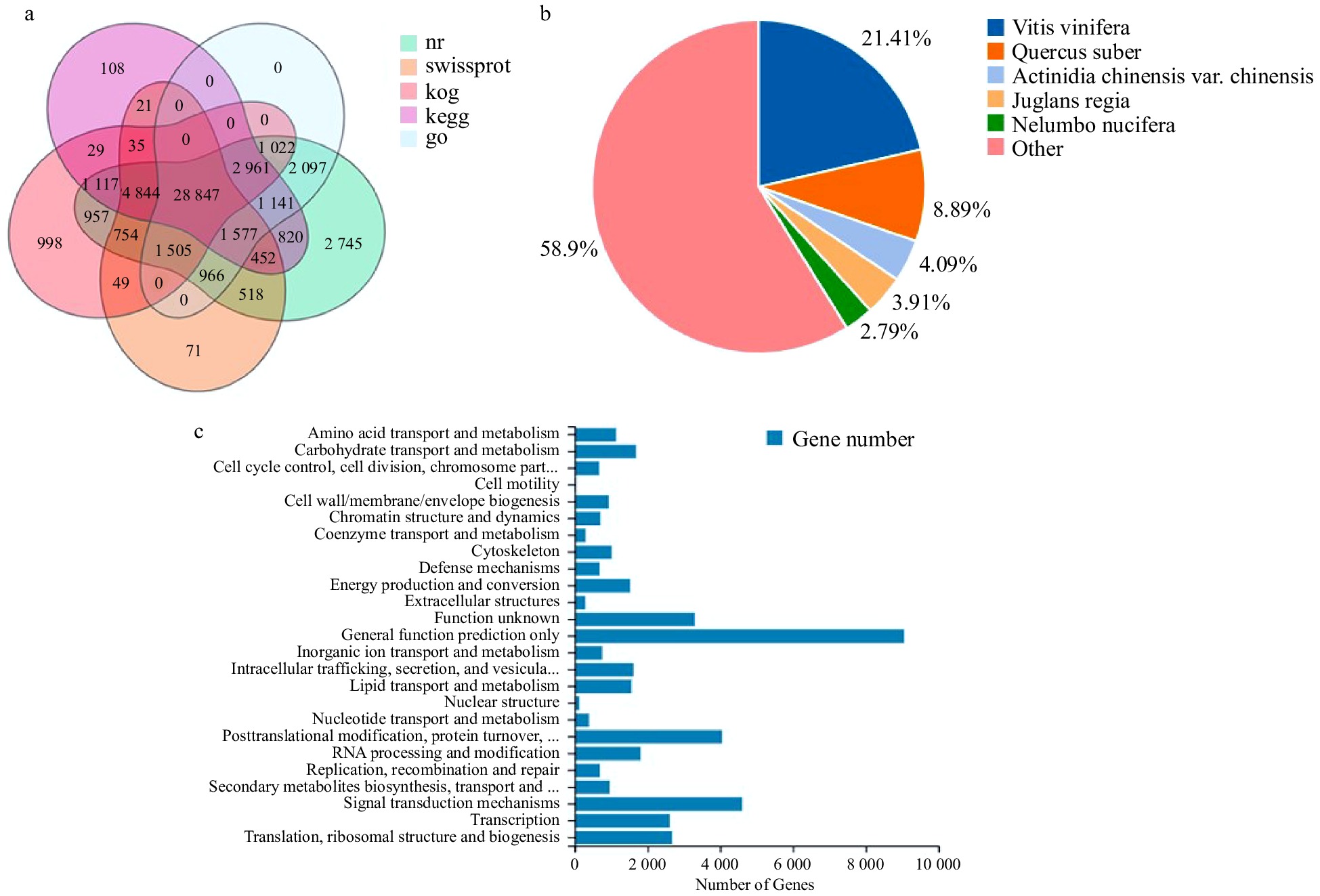
Figure 2. Gene functional annotation. (a) Venn diagram of the number of unigenes annotated in different public databases. (b) Species distribution of NR annotation. (c) Functional classification of KOG annotation. The x-axis represents the corresponding number of Unigene, the y-axis represents the KOG function classification name.
-
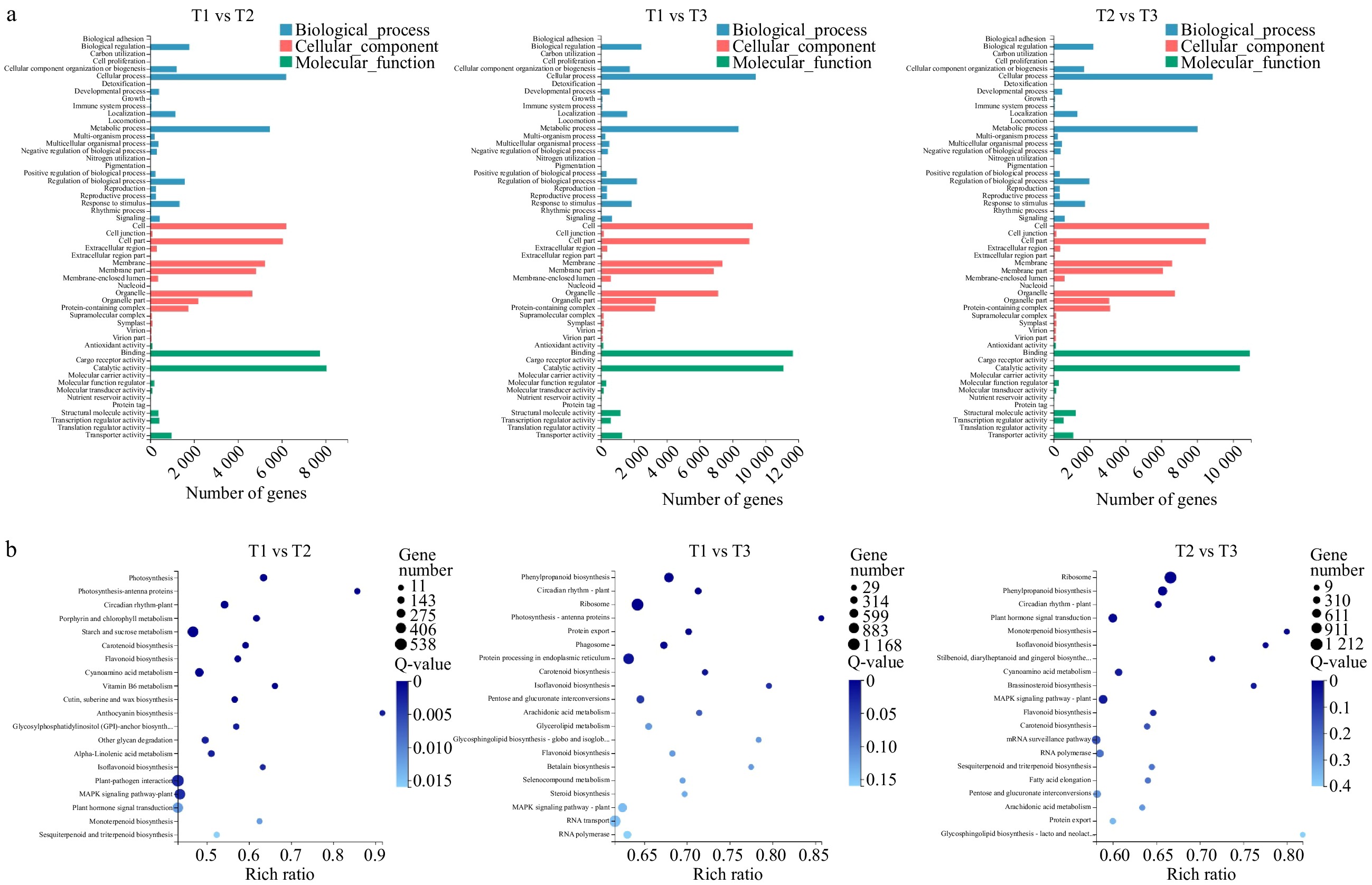
Figure 3. GO and KEGG Pathway analysis of DEGs. (a) Functional distribution of DEGs annotated GO. The x-axis represents the number of genes annotated GO and the y-axis represents the functional distribution of GO. (b) Bubble diagram of enrichment of DEGs KEGG pathway. The x-axis is enrichment ratio, the y-axis is KEGG Pathway, the bubble size indicates the number of genes annotated on one KEGG pathway, the color represents enrichment Q-value, and darker color represents smaller Q-value.
-
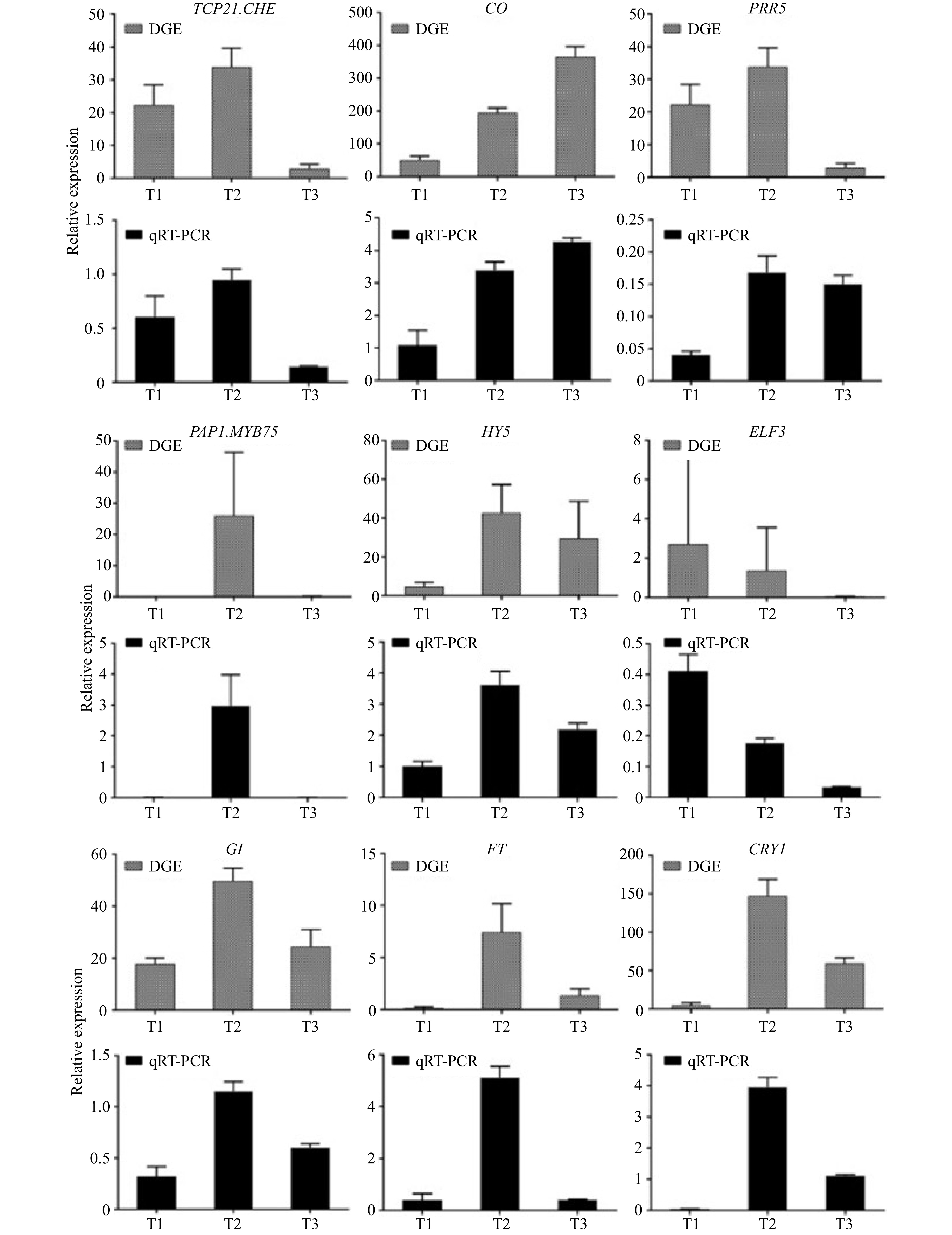
Figure 4. qRT-PCR validations of expression levels of DEGs.
-
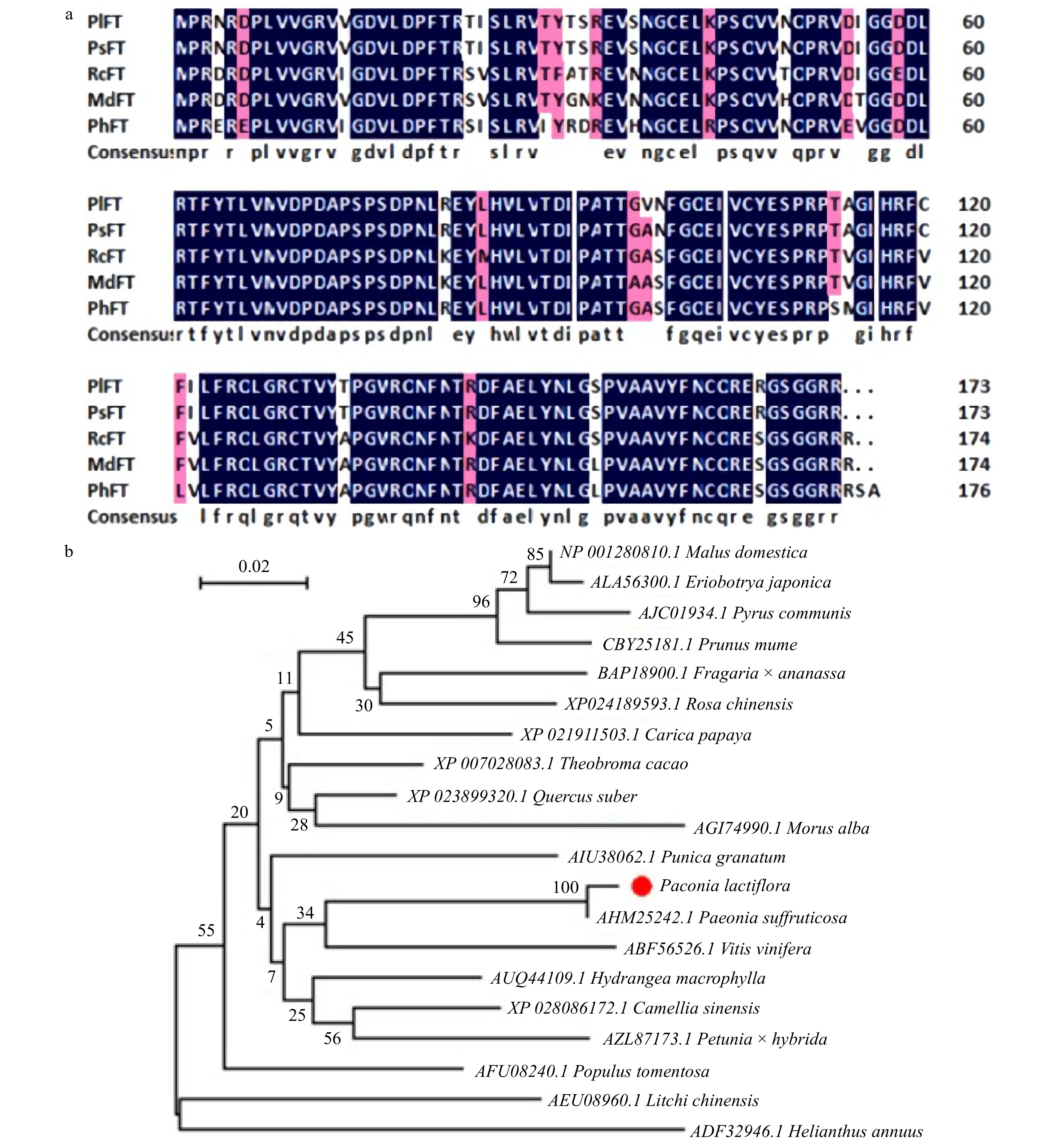
Figure 5. Sequence analysis of PlFT. (a) Amino-acid comparision between PlFT and FT homologues from other species. (b) Phylogenetic tree based on the amino acid sequences from PlFT and other species. AHM25242.1 [Paeonia suffruticosa]; AFU08240.1 [Populus tomentosa]; BAP18900.1 [Fragaria × ananassa]; AUQ44109.1 [Hydrangea macrophylla]; XP_023899320.1 [Quercus suber]; XP_028086172.1 [Camellia sinensis]; AIU38062.1 [Punica granatum]; XP_007028083.1 [Theobroma cacao]; XP_024189593.1 [Rosa chinensis]; AGI74990.1 [Morus alba]; XP_021911503.1 [Carica papaya]; ABF56526.1 [Vitis vinifera]; NP_001280810.1 [Malus domestica]; AZL87173.1 [Petunia × hybrida]; ALA56300.1 [Eriobotrya japonica]; AJC01934.1 [Pyrus communis]; AEU08960.1 [Litchi chinensis]; CBY25181.1 [Prunus mume]; ADF32946.1 [Helianthus annuus].
-
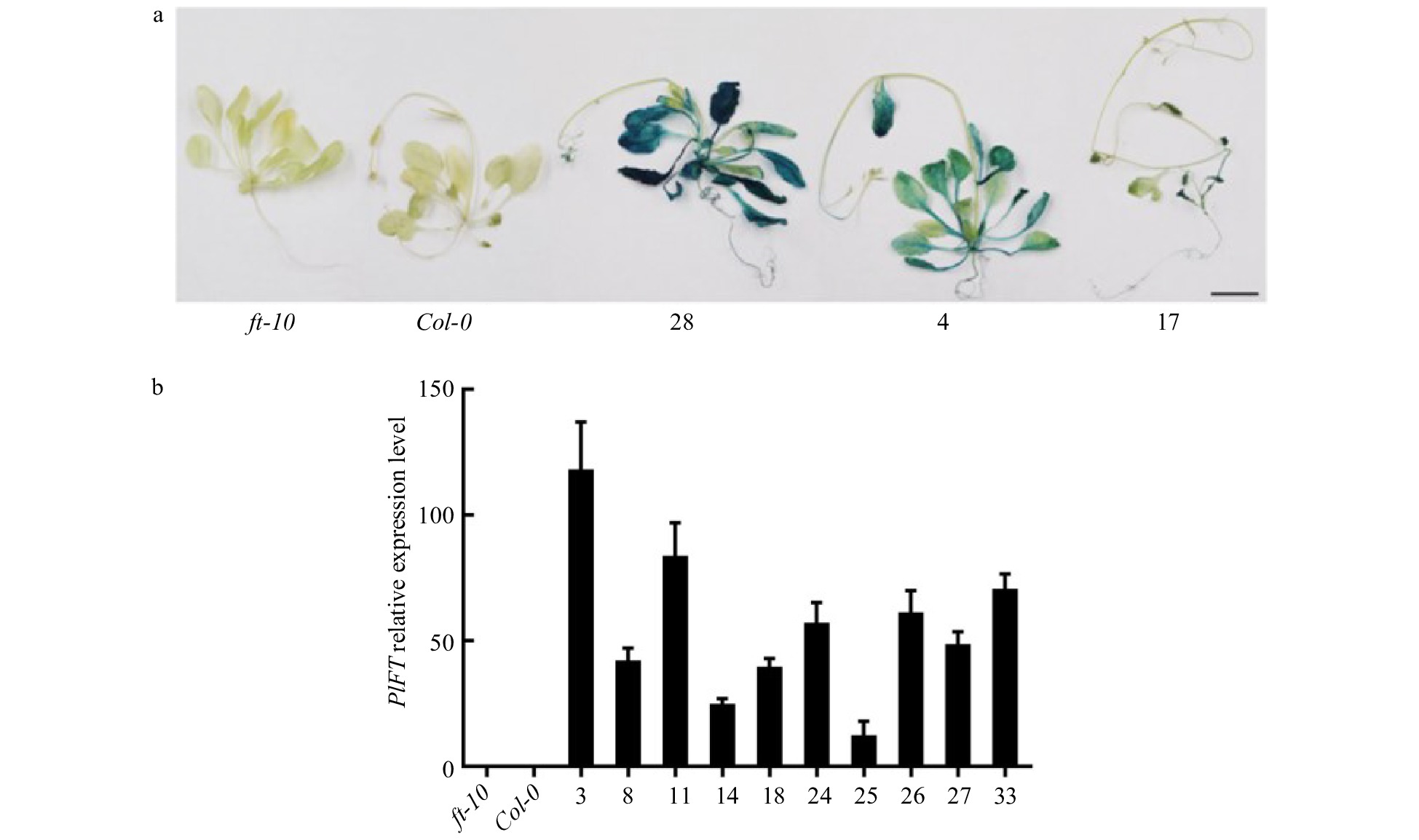
Figure 6. Identification of the PlFT gene in A. thaliana. (a) GUS staining results of transgenic Arabidopsis lines with PlFT gene. (b) Relative expression level of target genes in transgenic lines. ft-10 is mutant plant, Col-0 is wild-type plant, other numbers are PlFT transgenic plants.
-
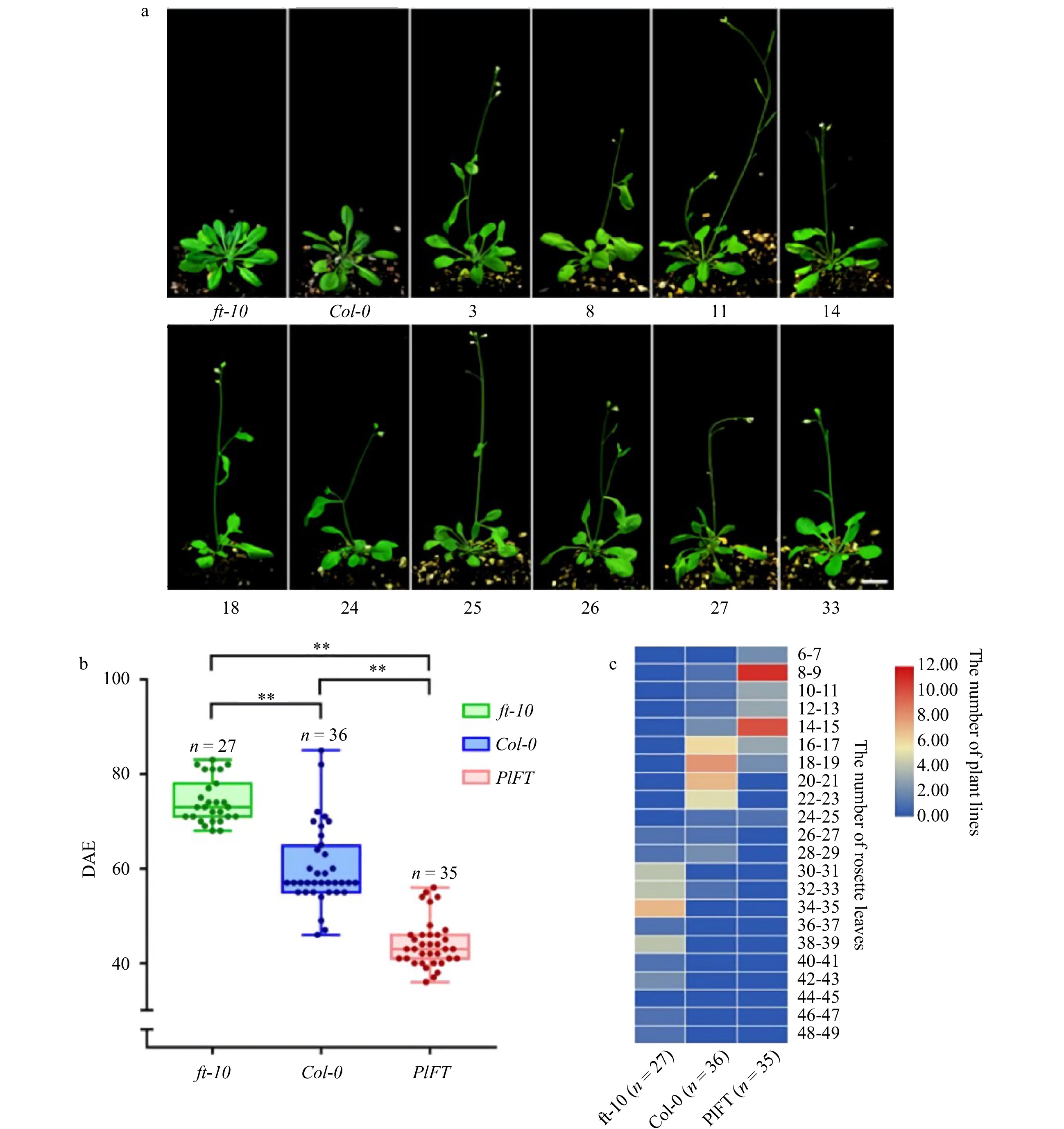
Figure 7. Phenotypic analysis of the PlFT gene in A. thaliana. (a) Phenotype of transgenic Arabidopsis (Scale = 1 cm). (b) Flowering time of Arabidopsis DAE in the figure indicates the days after emergence and indicate significant differences (P < 0.01). (c) Heat map of the leaves of the rosette of Arabidopsis thaliana.
-
Gene ID Entry Name log2(T2/T1) log2(T3/T1) log2(T3/T2) T1 vs T2 T1 vs T3 T2 vs T3 CL1090.Contig1_All K16221 TCP21, CHE −1.22 −1.88 down down CL1090.Contig2_All −1.70 −2.21 down down Unigene12923_All −3.09 −2.39 down down Unigene15030_All 2.53 2.33 up up Unigene17026_All −1.13 down Unigene19482_All −3.00 −3.60 down down Unigene6346_All 2.27 1.44 up up CL1148.Contig1_All K12121 PHYB 4.43 11.10 6.68 up up up CL1148.Contig4_All 1.58 7.06 5.48 up up up CL1516.Contig10_All 2.70 7.40 4.70 up up up CL1516.Contig11_All 4.12 8.90 4.78 up up up CL1516.Contig2_All 2.97 1.25 −1.72 up up down CL1516.Contig3_All 5.19 9.32 4.12 up up up CL1516.Contig4_All 6.56 6.69 up up CL1516.Contig5_All −2.89 −6.12 down down CL1516.Contig6_All −8.14 −8.16 down down CL1516.Contig7_All −3.44 5.73 9.17 down up up CL1516.Contig8_All 1.11 −1.54 −2.65 up down down CL1516.Contig9_All 1.84 5.98 4.13 up up up CL175.Contig2_All 2.40 2.36 up up CL175.Contig4_All 2.26 1.91 up up CL5438.Contig1_All −1.79 −8.85 −7.07 down down down CL6053.Contig1_All 1.19 up CL6053.Contig2_All 2.58 4.17 1.59 up up up CL6053.Contig3_All −1.34 1.94 3.28 down up up CL6068.Contig1_All 4.60 4.51 up up CL6900.Contig1_All 1.30 −7.36 −8.65 up down down CL6900.Contig2_All 1.09 1.36 up up CL7074.Contig1_All 8.14 8.04 up up CL7690.Contig3_All −1.93 −2.80 down down CL7690.Contig4_All 1.01 −7.93 −8.94 up down down CL7690.Contig5_All 1.54 up Unigene12459_All 1.26 −1.04 up down Unigene12460_All 6.55 6.45 up up Unigene12461_All 3.70 8.02 4.32 up up up Unigene1351_All 9.45 9.35 up up Unigene15547_All 9.58 9.48 up up Unigene24958_All −4.59 2.60 7.20 down up up Unigene25706_All 8.96 7.86 up up Unigene39640_All −1.07 down Unigene6288_All 1.24 up Unigene6289_All 1.29 up CL1150.Contig3_All K12133 LHY 3.80 3.14 up up CL1150.Contig4_All 3.98 4.05 up up CL1150.Contig5_All 6.15 6.25 up up CL1150.Contig7_All 4.35 4.23 up up CL1184.Contig1_All 2.71 −2.01 up down CL5076.Contig1_All −1.34 down CL5076.Contig3_All −1.62 down CL5076.Contig4_All −2.16 −7.47 −5.31 down down down CL5076.Contig5_All −1.21 down CL8141.Contig1_All −1.09 2.51 3.60 down up up CL8141.Contig2_All 2.33 2.79 up up CL1267.Contig1_All K12135 CO 1.16 2.88 1.72 up up up CL1672.Contig2_All 1.13 up CL3591.Contig1_All 1.59 −1.09 −2.68 up down down CL3591.Contig2_All 2.67 −2.06 −4.73 up down down CL3591.Contig3_All 2.35 −6.78 −9.13 up down down CL3591.Contig4_All 3.10 −1.09 −4.19 up down down CL3591.Contig5_All −1.60 −2.47 down down CL409.Contig3_All −1.11 −1.56 down down CL409.Contig4_All −1.39 down CL409.Contig5_All 1.21 −1.52 up down CL4357.Contig1_All 7.30 8.69 1.38 up up up CL4357.Contig2_All 4.39 3.23 −1.16 up up down CL4357.Contig3_All 9.23 11.51 2.28 up up up CL4357.Contig4_All 2.96 4.33 1.37 up up up CL4357.Contig5_All 5.08 1.55 −3.52 up up down CL6692.Contig3_All 2.25 −4.50 −6.75 up down down CL7718.Contig1_All 1.65 −1.49 −3.14 up down down CL7718.Contig2_All 1.44 −1.29 up down CL7917.Contig1_All −1.38 down CL7917.Contig2_All −1.28 −1.01 down down CL7917.Contig3_All −1.75 −1.79 down down CL7932.Contig1_All 1.92 2.14 up up CL7932.Contig2_All 1.95 2.86 up up Unigene11310_All 2.14 −2.24 up down Unigene1286_All 1.84 1.25 up up Unigene14242_All 3.42 7.71 4.29 up up up Unigene17669_All 6.48 4.58 −1.90 up up down Unigene32926_All 4.14 −5.07 up down Unigene410_All 5.11 5.13 up up Unigene496_All 2.47 −2.83 up down Unigene9300_All 1.86 7.00 5.14 up up up CL3891.Contig1_All K12130 PRR5 7.17 4.47 −2.70 up up down CL3891.Contig2_All 3.36 −7.07 up down CL3891.Contig3_All 2.69 −2.64 up down CL3891.Contig4_All 2.56 −2.46 up down CL6193.Contig1_All 4.75 3.70 −1.05 up up down CL6193.Contig2_All 4.31 2.52 −1.78 up up down Unigene8183_All 2.55 −2.91 up down Unigene8184_All 3.33 −2.79 up down CL4229.Contig1_All K12129 PRR7 1.21 2.86 1.65 up up up CL4229.Contig10_All 1.44 3.38 1.94 up up up CL4229.Contig2_All 1.59 3.96 2.38 up up up CL4229.Contig3_All 1.46 2.59 1.13 up up up CL4229.Contig4_All 1.83 1.78 up up CL4229.Contig5_All 4.09 4.59 up up CL4229.Contig6_All 3.02 −6.05 −9.07 up down down CL4229.Contig7_All 3.20 6.69 3.48 up up up CL4229.Contig8_All 2.51 2.28 up up CL4229.Contig9_All 6.17 7.13 up up CL7872.Contig1_All 1.32 2.56 1.24 up up up CL7872.Contig2_All 2.34 3.67 1.33 up up up Unigene6980_All 4.16 2.26 −1.90 up up down CL4436.Contig1_All K12116 FKF1 −1.34 −1.58 down down CL4436.Contig2_All 1.13 −1.20 up down CL4719.Contig1_All K16222 CDF1 −1.27 1.27 down up CL5753.Contig1_All 3.95 2.85 down down CL5753.Contig2_All −2.77 −3.64 down down down CL5837.Contig1_All −1.83 −2.67 up up up CL5837.Contig2_All −3.49 −5.83 up up up CL6631.Contig2_All −3.77 −3.58 down down CL8648.Contig1_All −1.90 down down down CL8648.Contig2_All −1.36 down down down Unigene10801_All 10.91 −7.84 up up Unigene11044_All −2.71 −8.05 −5.34 up Unigene12006_All 7.60 5.50 up down Unigene12046_All −2.30 −1.32 up up up Unigene12573_All −2.04 −4.46 −2.43 down down up Unigene14940_All 7.05 10.78 3.73 up down Unigene14950_All 7.24 13.54 6.30 down down down Unigene695_All −2.86 −2.83 up up Unigene972_All −2.18 −6.64 −4.47 up up Unigene15046_All −2.45 −5.86 −3.41 up down CL5158.Contig1_All K12120 PHYA 1.44 1.08 up up CL9769.Contig1_All 1.65 down down CL9769.Contig2_All 1.95 −1.86 down down CL9769.Contig3_All 4.27 5.83 1.56 down down Unigene14810_All −3.09 −1.66 1.43 down down Unigene2317_All 2.03 −2.02 down Unigene2317_All −3.39 −5.28 −1.89 down CL5739.Contig1_All K16166 PAP1, MYB75 1.94 2.48 up down CL5739.Contig3_All 1.89 1.64 down down down Unigene15771_All 1.81 −1.74 up up CL6391.Contig1_All K16241 HY5 3.25 2.69 up up CL6391.Contig2_All 4.25 1.38 −2.87 up up down CL6391.Contig3_All 5.39 −4.60 up down CL9659.Contig1_All −1.22 −1.90 down down Unigene6996_All 8.00 7.61 up up CL6767.Contig1_All K12127 TOC1, APRR1 2.28 1.49 up up CL6767.Contig2_All −1.25 −1.37 down down CL6767.Contig3_All −2.52 −3.05 down down CL835.Contig1_All 2.93 −2.80 −5.73 up down down CL835.Contig2_All −3.37 −3.31 down down CL835.Contig3_All −2.37 −1.71 down down CL835.Contig4_All −5.89 −5.70 down down CL835.Contig5_All 2.05 −3.30 −5.35 up down down CL835.Contig6_All −3.41 −3.94 down down CL835.Contig7_All 1.25 −2.26 −3.51 up down down Unigene15499_All 6.55 1.10 −5.45 up up down CL7407.Contig1_All K12125 ELF3 −6.46 −5.48 down down CL8526.Contig1_All K12124 GI 1.48 −1.02 up down CL8526.Contig2_All 2.59 −3.56 −6.16 up down down CL8526.Contig3_All 1.65 1.11 up up CL8526.Contig4_All 1.51 up CL8783.Contig1_All K16223 FT 9.27 6.48 −2.78 up up down CL8783.Contig2_All 6.12 3.64 −2.48 up up down CL8783.Contig3_All 7.57 5.02 −2.55 up up down Unigene18266_All −5.81 −4.83 down down Unigene23647_All −5.49 −2.51 down down Unigene30306_All 7.14 7.05 up up Unigene35531_All −8.42 −8.44 down down Unigene36039_All −3.55 −7.15 down down Unigene235_All K12118 CRY1 5.47 4.40 −1.07 up up down Unigene236_All 5.01 3.70 −1.31 up up down CL926.Contig1_All K12119 CRY2 2.81 2.28 up up CL926.Contig11_All 3.01 2.38 up up CL926.Contig2_All 2.37 3.07 up up CL926.Contig3_All 1.71 1.69 up up CL926.Contig4_All 1.12 2.06 up up CL926.Contig5_All 1.04 3.57 2.53 up up up CL926.Contig6_All 1.22 5.43 4.20 up up up CL926.Contig7_All 2.79 2.28 up up CL926.Contig8_All 2.09 5.79 3.70 up up up Unigene2881_All −1.41 1.19 2.60 down up up Unigene21537_All K12115 ZTL 1.57 up Unigene32913_All 1.25 up Unigene8078_All −1.24 1.58 down up Unigene8082_All 1.96 up Unigene8325_All −1.95 −2.92 down down Unigene6219_All K16240 SPA1 2.77 1.31 −1.45 up up down CL773.Contig1_All K01184 FLC 3.09 −6.44 −9.54 up down down CL773.Contig3_All 4.41 −4.89 −9.29 up down down CL773.Contig4_All 4.24 −4.90 −9.14 up down down CL773.Contig5_All 3.79 −6.37 −10.16 up down down Unigene16702_All 3.22 −6.84 −10.07 up down down Unigene16704_All −4.77 −5.08 down down CL7416.Contig1_All K09264 FUL 3.39 3.44 up up CL7416.Contig2_All 3.51 3.34 up up Unigene18421_All 4.66 −4.95 up down Unigene44881_All 5.31 3.57 −1.75 up up down Unigene44883_All 4.71 −7.96 up down Unigene7311_All K09264 AP1 7.44 12.69 5.25 up up up CL8905.Contig1_All K09260 SOC1 2.35 −3.64 −5.99 up down down Unigene7203_All −3.22 −7.68 −4.46 down down down Table 1. Analysis of main DEGs for KEGG pathway related with flowering of 'Dafugui' samples in different periods.
Figures
(7)
Tables
(1)