-

Figure 1. (a) COG function annotation of stigma transcriptome. (b) GO function annotation of stigma transcriptome. (c) COG function annotation of anther transcriptome. (d) GO function annotation of anther transcriptome.
-
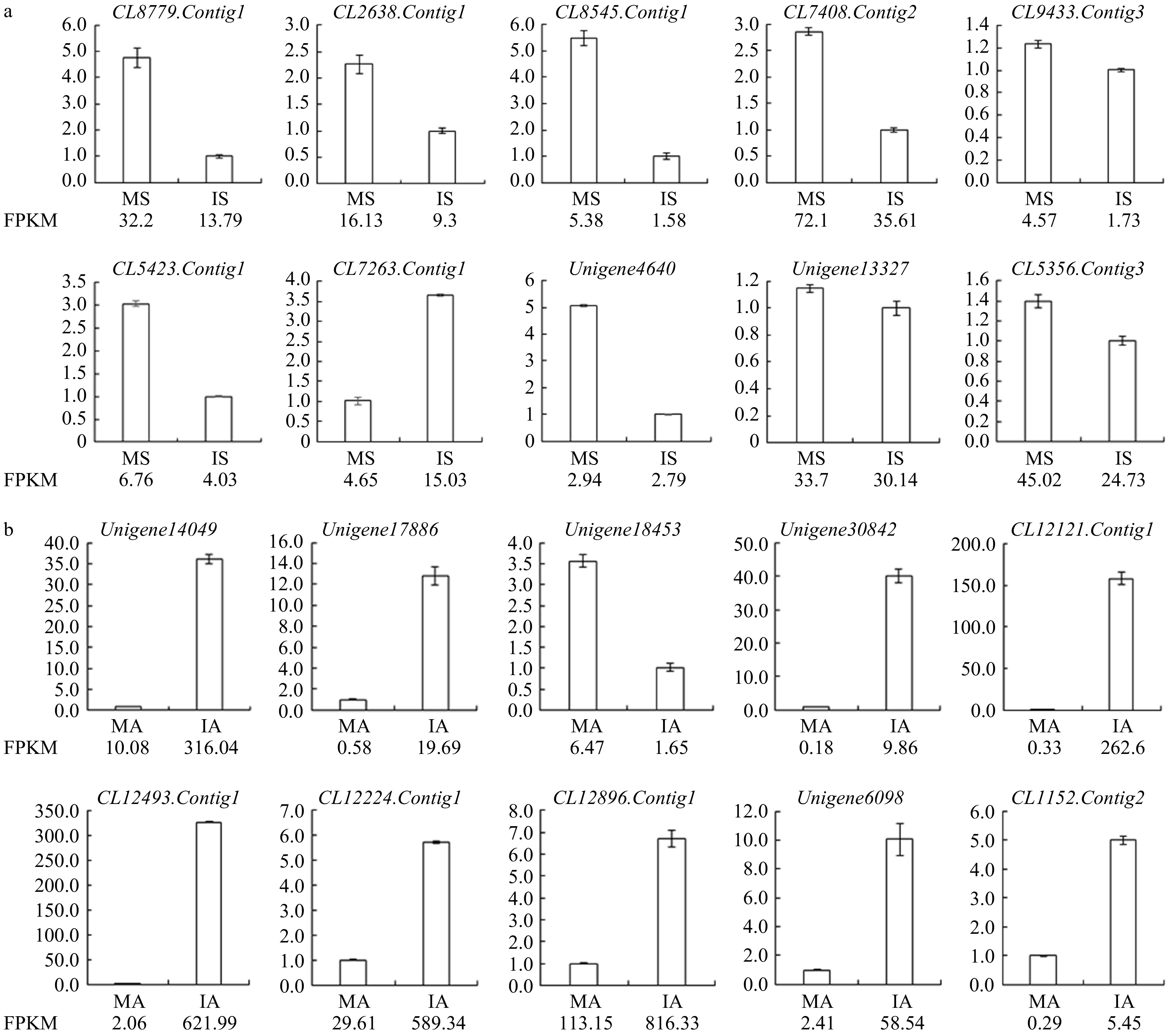
Figure 2. (a) qRT-PCR verification of RNA-Seq results of stigma transcriptome. (b) qRT-PCR verification of RNA-Seq results of anther transcriptome.
-
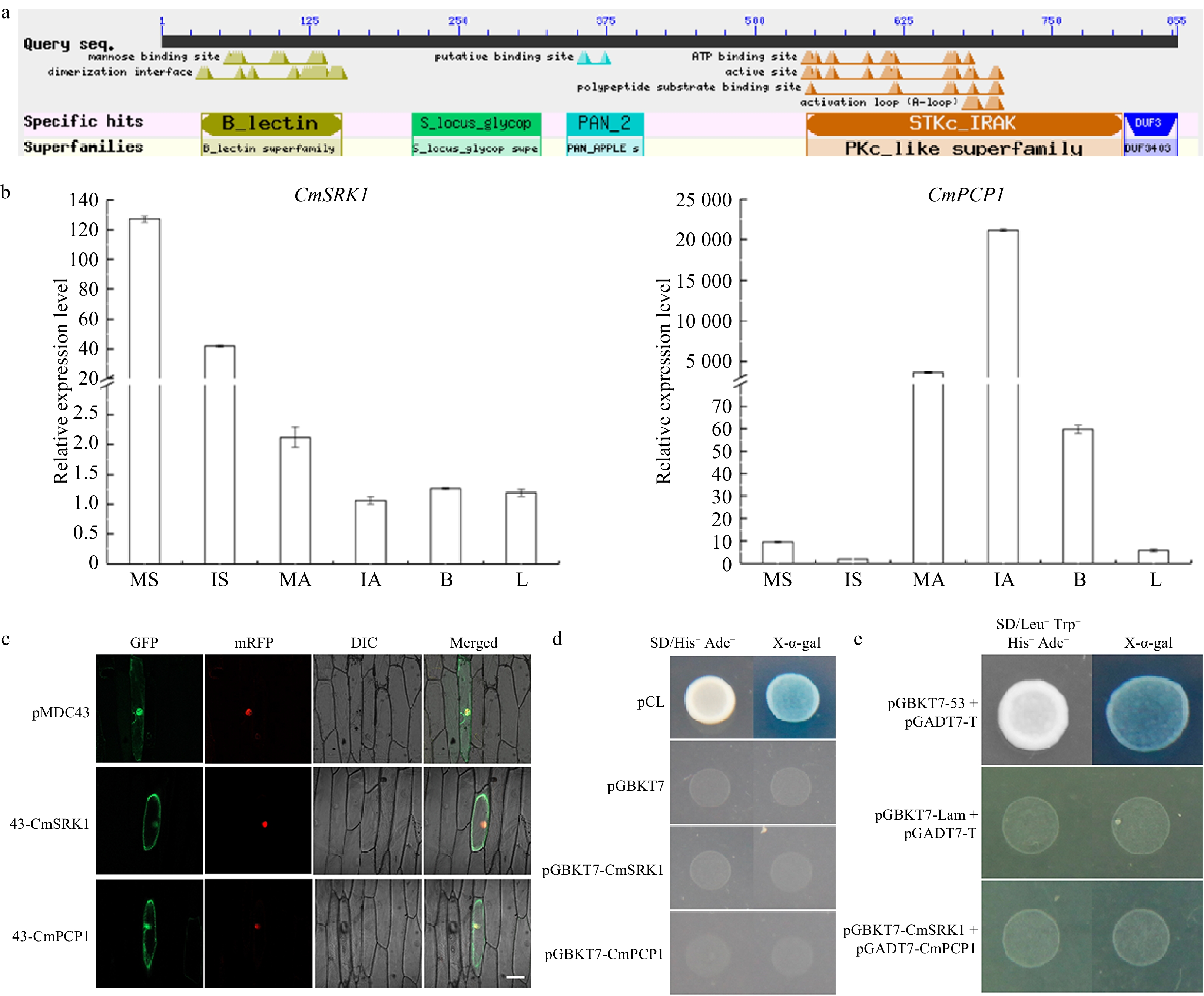
Figure 3. (a) Blast result of CmSRK1. (b) Expression profiles of CmSRK1 and CmPCP1 in different tissues of 'Q10-22-2'. MS: mature stigmas; IS: immature stigmas; MA: mature anthers; IA: immature anthers; B: flower buds; L: leaves. (c) Subcellular localization of CmSRK1 and CmPCP1. Bar=100 μm. (d) Transcriptional activation activity of CmSRK1 and CmPCP1. (e) Yeast two hybrid between CmSRK1 and CmPCP1.
-
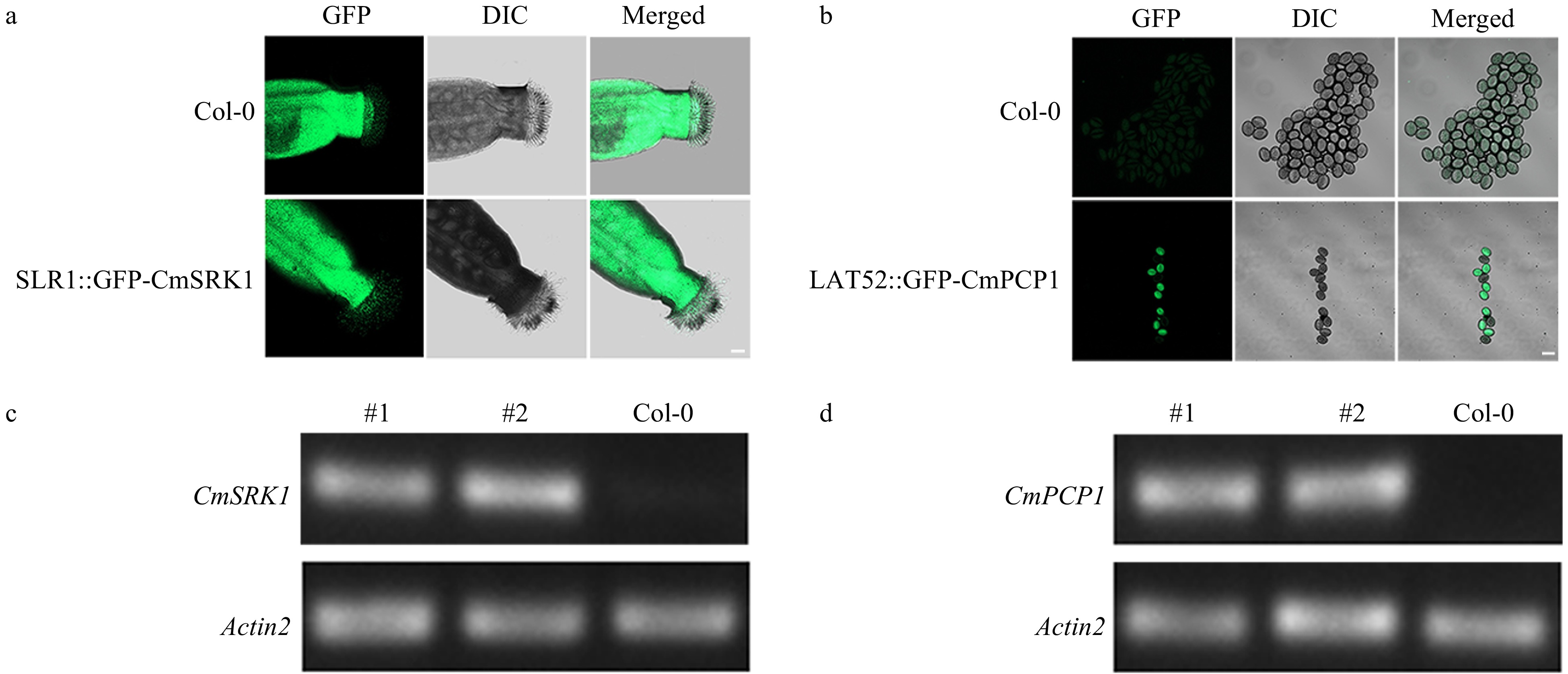
Figure 4. a,b GFP signals in stigmas and pollens of Arabidopsis. (a) GFP signal in stigmas of Arabidopsis; (b) GFP signals in pollens of Arabidopsis. Bar = 20 μm. (c) RT-PCR identification of CmSRK1 in Arabidopsis; (d) RT-PCR identification of CmPCP1 in Arabidopsis. #1 and #2: two transgenic lines.
-
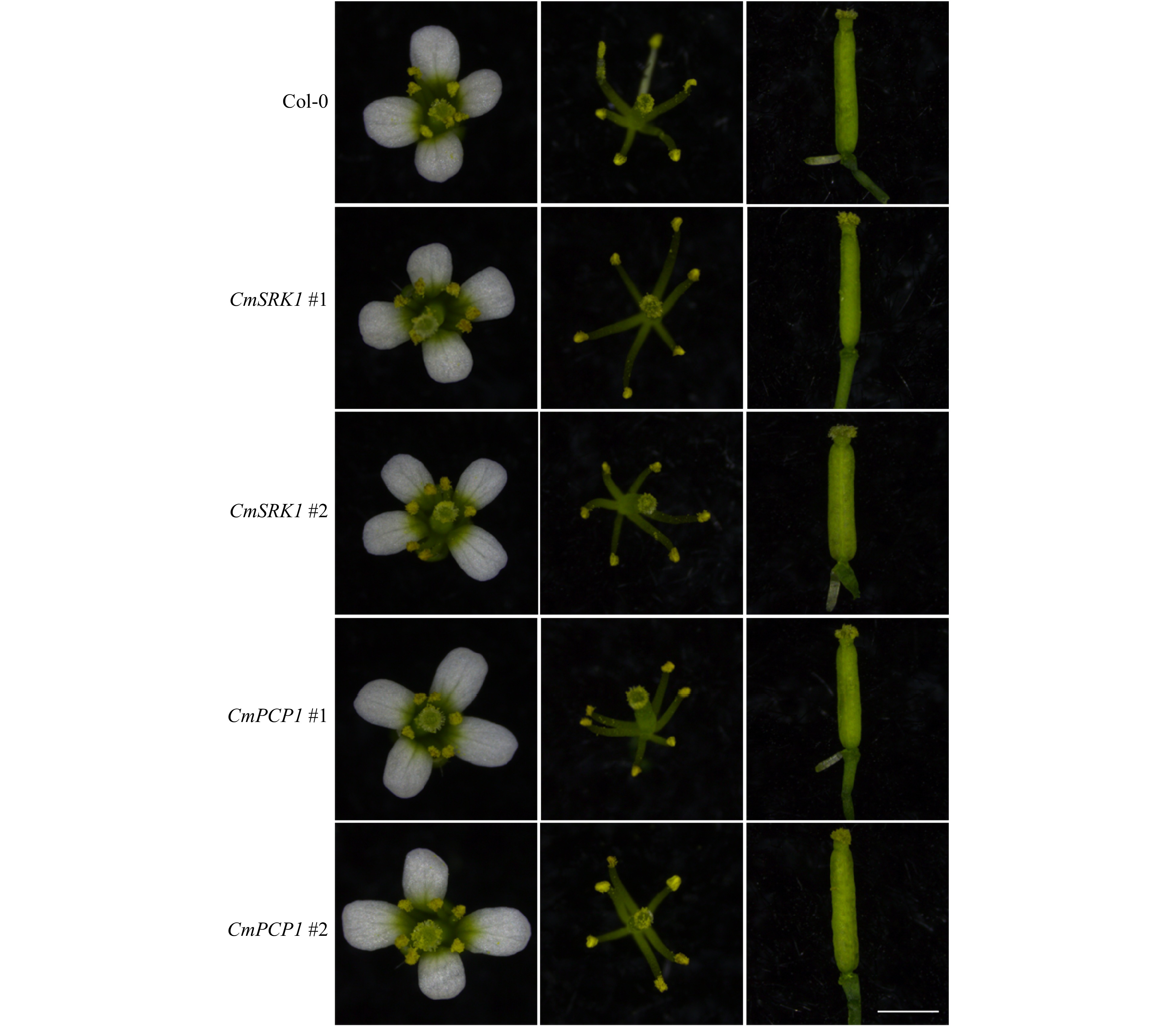
Figure 5. Floral organ observation of Arabidopsis after transformation of CmSRK1 and CmPCP1. The first column was the whole flowers, the second was the pistils and stamens, and the third column was the pistils. The arabidopsis used for observation was 30 d after planting. Bar = 1 mm.
-
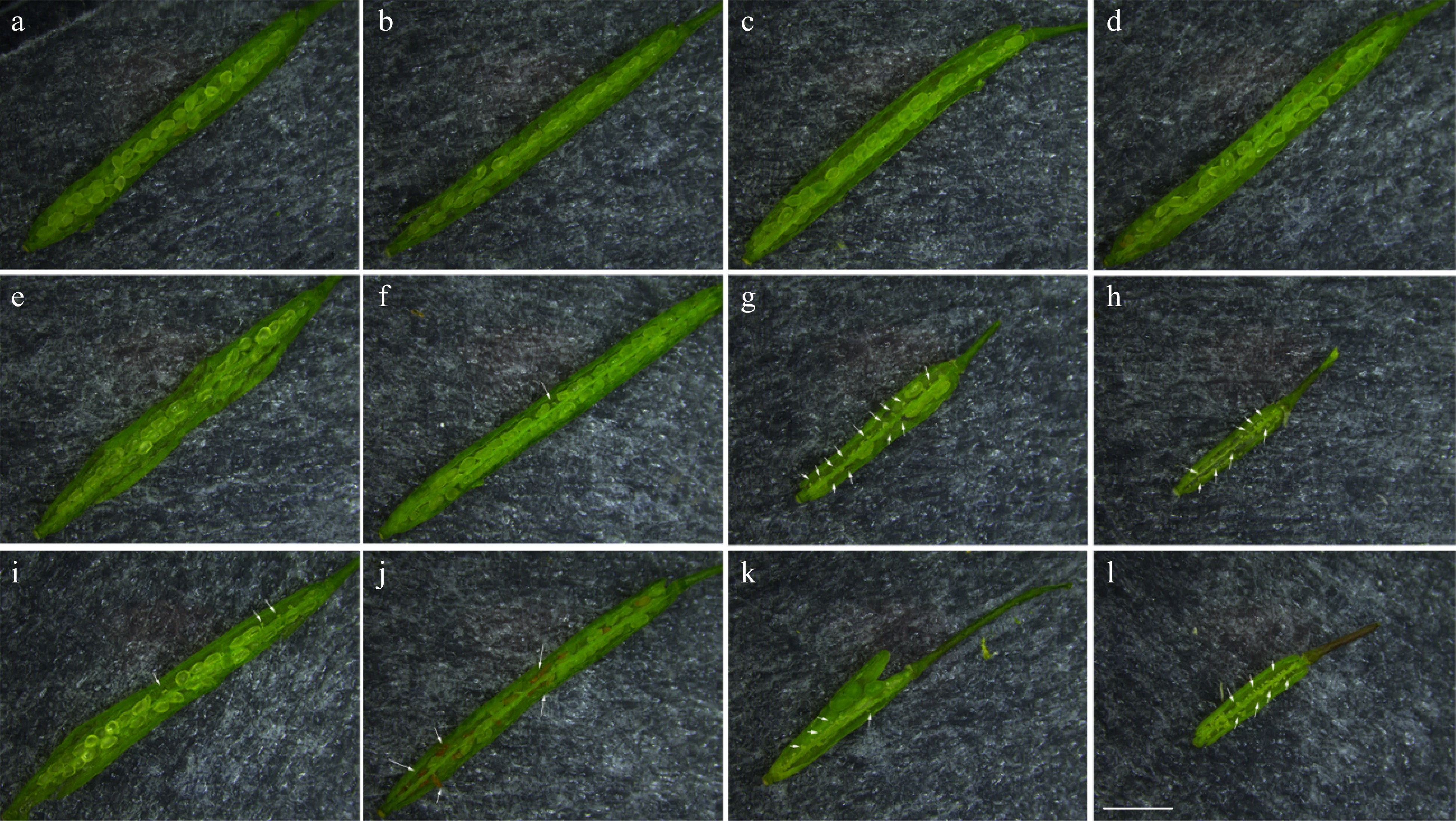
Figure 6. Anatomical observation of self-pollinated and cross-pollinated fruit pods of Arabidopsis. (a, b) Col-0⊗; (c, d) Col-0 × Col-0; (e) CmSRK1 #1⊗; (f) CmPCP1 #1⊗; (g, h) CmSRK1 #1 × CmPCP1 #1; (i) CmSRK1 #2⊗; (j) CmPCP1 #2⊗; (k, l) CmSRK1 #2 × CmPCP1 #2. The white arrows identify the defected position in transgenic Arabidopsis. Bar = 2 mm.
-
Unigene ID MS-FPKM IS-FPKM Annotation CL7408.Contig2 72.1 35.61 S-receptor-like serine/threonine-protein kinase RLK1 CL5423.Contig1 6.76 4.03 S-receptor-like serine/threonine-protein kinase At1g11410 Unigene50549 5.71 0 S-receptor-like serine/threonine-protein kinase SD1-1 CL6070.Contig3 5.43 4.42 S-receptor-like serine/threonine-protein kinase At4g27290 CL8545.Contig1 5.38 1.58 S-receptor-like serine/threonine-protein kinase SD1-1 CL9433.Contig3 4.57 1.73 S-receptor-like serine/threonine-protein kinase At2g19130 Unigene17909 3.42 1.6 S-receptor-like serine/threonine-protein kinase At1g34300 Unigene10651 3.37 1.36 S-receptor-like serine/threonine-protein kinase SD3-1 CL1678.Contig13 3.21 1.1 S-receptor-like serine/threonine-protein kinase B120 CL1678.Contig3 3.18 1.49 S-receptor-like serine/threonine-protein kinase At1g67520 Unigene54285 2.97 0 S-receptor-like serine/threonine-protein kinase At4g27290 CL6328.Contig1 2.9 1.25 S-receptor-like serine/threonine-protein kinase RLK1 Unigene5472 2.46 0.82 S-receptor-like serine/threonine-protein kinase At5g24080 Table 1. Candidate pistil S genes in 'Q10-22-2's stigmas.
-
Unigene ID MA-FPKM IA-FPKM Annotation CL12121.Contig1 0.33 262.6 S locus-related glycoprotein 1 binding pollen coat protein CL12224.Contig1 29.61 589.34 pollen coat-like protein CL12224.Contig2 12.22 156.65 pollen coat-like protein CL12493.Contig1 2.06 621.99 S locus-related glycoprotein 1 binding pollen coat protein CL12896.Contig1 113.15 816.33 pollen coat-like protein Table 2. Candidate pollen S genes in 'Q10-22-2's anthers.
-
Self-pollination/cross-pollination Seed set (%) Col-0 93.58 ± 2.17a Col-0 × Col-0 84.43 ± 5.01a CmSRK1 #1 84.89 ± 3.85a CmPCP1 #1 94.87 ± 2.25a CmSRK1 #1 × CmPCP1 #1 19.62 ± 6.70c CmSRK1 #2 78.26 ± 7.12a CmPCP1 #2 50.51 ± 6.60b CmSRK1 #2 × CmPCP1 #2 11.64 ± 3.68c Values given were mean ± standard error. Values with different superscript indicated significant differences at p ≤ 0.05 according to Tukey’s test. Table 3. Self-pollinated and cross-pollinated seed sets of Arabidopsis.
Figures
(6)
Tables
(3)