-
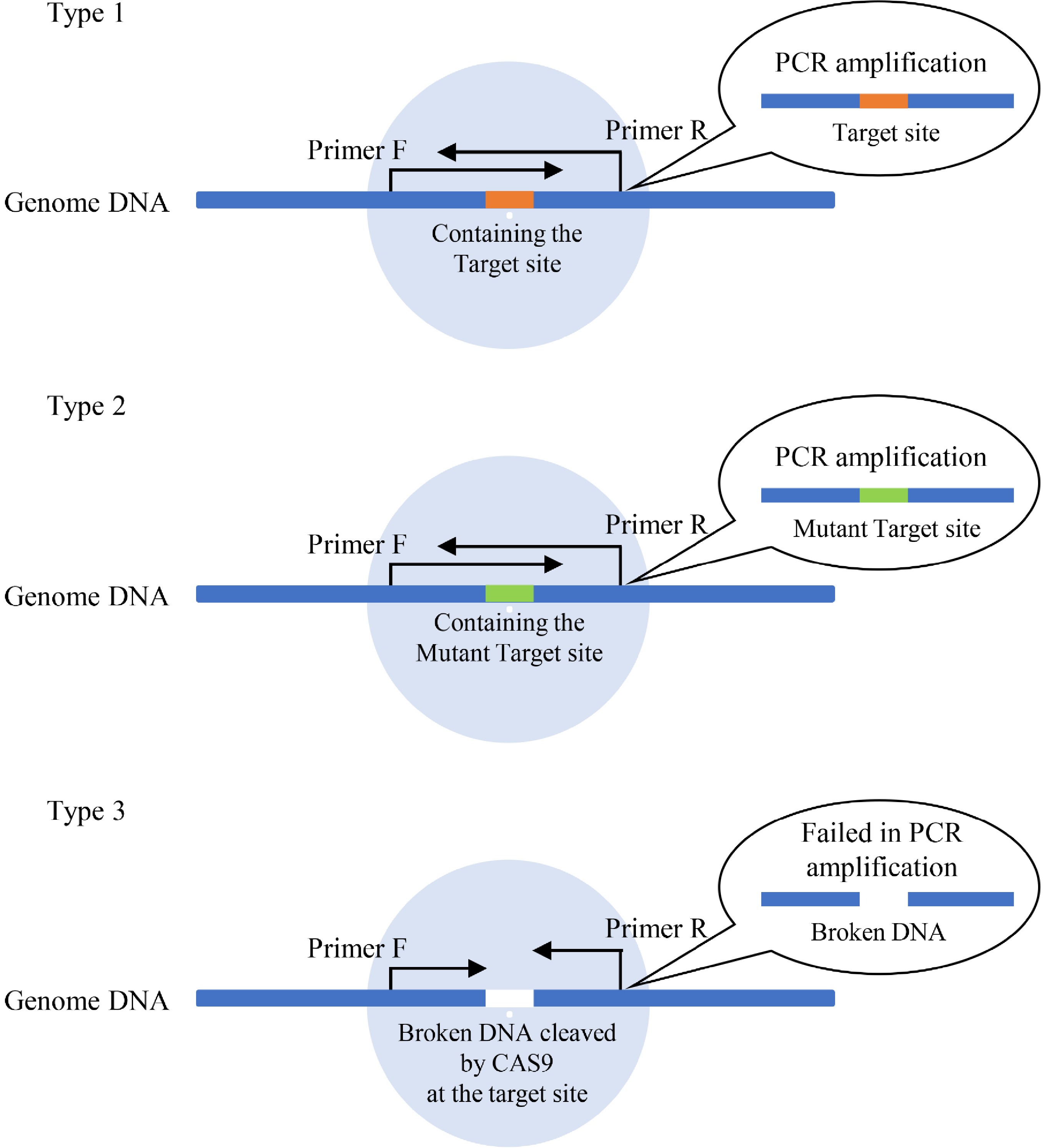
Figure 1. The principle of determination of efficiencies of Cas9 cutting. There are 3 types of DNA present in plant cells after transient transformation of gRNA and Cas9. Type 1: DNA containing the target Cas9 cutting site without breakage (which can be PCR amplified). This kind of DNA has not been cleaved by Cas9 or has been cleaved by Cas9 but has been repaired without a mutant. Type 2: DNA containing the mutant target CRISPR cutting site without breakage (which can be PCR amplified). This kind of DNA had been cut by Cas9 and repaired with mutation. Type 3: the broken DNA by cutting on the target site by CRISPR/Cas. Type 3 DNA cannot be PCR amplified, and its quantities can reflect the efficiency of Cas9 cutting and be detected by qPCR.
-
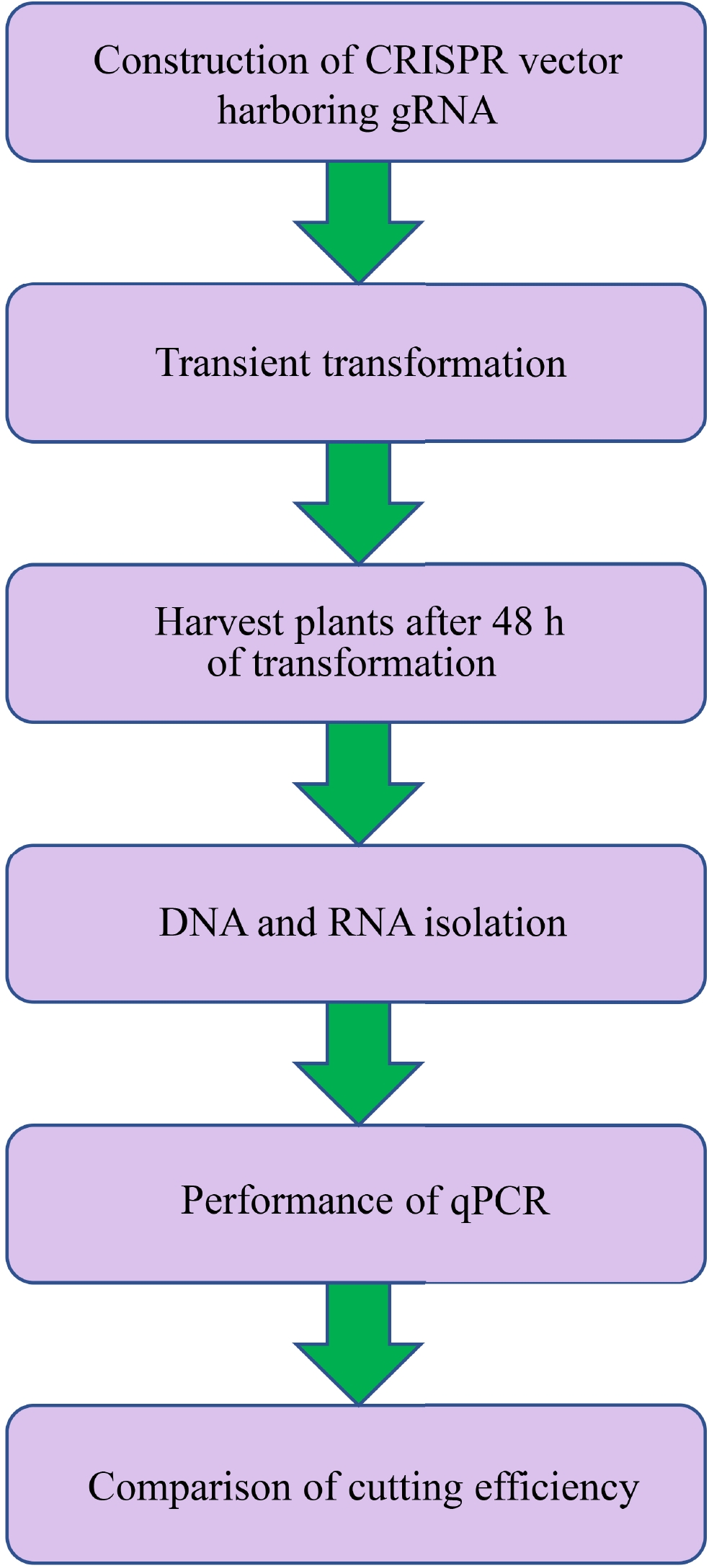
Figure 2. Flowchart of transient CRISPR/Cas editing in plants (TCEP). (1) CRISPR/Cas vectors harboring gRNAs were constructed and transformed into Agrobacterium for plant transformation. (2) Transient transformation was performed on the plants. (3) After transformation for 48 h, the plants were harvested for DNA isolation. (4) Genomic DNA and total RNA were extracted, and RNA was reverse transcribed into cDNA. (5) Quantitative PCR was performed to determine the cutting efficiency, and the cutting efficiency was determined by the amount of broken DNA normalized by the expression of Cas9.
-

Figure 3. Determination of the efficiencies of Cas9 cutting to different target sites in birch. Five target sites were selected for study, and the efficiencies of Cas9 cutting to different target sites were studied using qPCR. (a): Determination of Cas9 cutting efficiency using the TCEP method in birch. (b): Determination of Cas9 cutting efficiency using the in vitro cutting method. Targets 1–5: The CRISPR target cutting sites 1–5 of birch and the sequences of these target sites are shown in Supplemental Table S1. The target site with the lowest cutting efficiency was set as 1 to normalize the efficiencies of Cas9 cutting to other target sites.
-
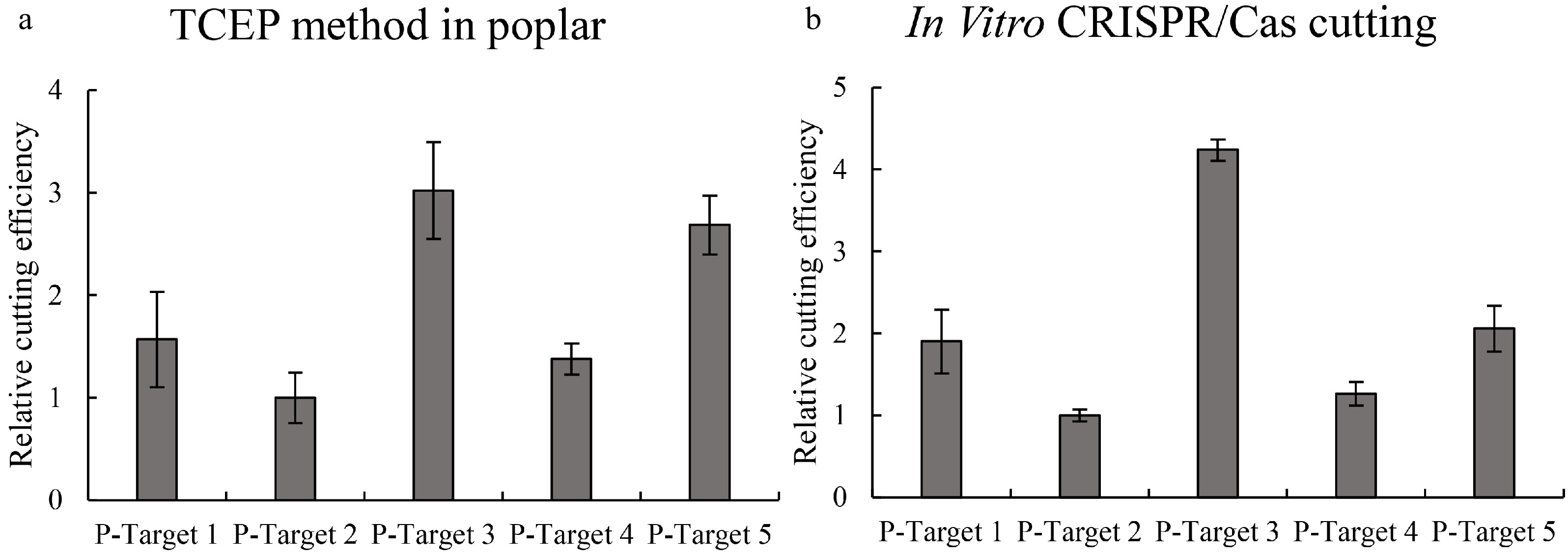
Figure 4. Determination of the efficiencies of Cas9 cutting to different target sites in poplar plants. Five target sites were studied, and the efficiencies of Cas9 cutting to different target sites were determined using qPCR. (a): Determination of Cas9 cutting efficiency using the TCEP method in poplar (P. davidiana×P. bolleana). (b): Determination of Cas9 cutting efficiency using the in vitro cutting method. Targets 1–5: CRISPR target cutting sites 1–5 of poplar, and the sequences of these target sites are shown in Supplemental Table S1. The target site with the lowest cutting efficiency was set as 1 to normalize the efficiencies of Cas9 cutting to other target sites. * indicates a significant difference compared with the control (p < 0.05).
-
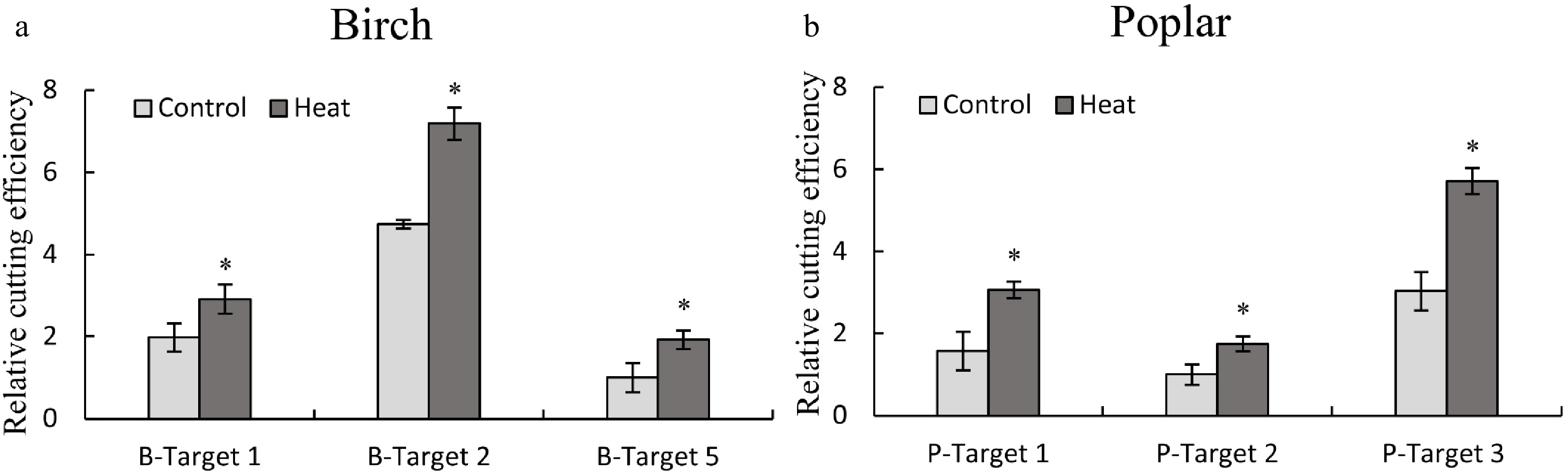
Figure 5. Determination of the effects of heat treatment on the cutting efficiency of CRISPR/Cas using the TCEP method. (a, b): The effects of heat treatment on CRISPR/Cas efficiency in birch (a) and poplar (b). Three target sites with high, medium and low CRISPR cutting efficiency were selected from birch and poplar for study using qPCR. After transient transformation for 48 h, the plants were incubated at 35 °C for 24 h (Heat treatment), and the plants without heat treatment were harvested at the same time as the control (Control). The serial numbers of target cutting sites of birch and poplar are consistent with Fig. 2 and Fig. 3, respectively. The target site with the lowest cutting efficiency was set as 1 to normalize the efficiencies of Cas9 cutting to other target sites. * indicates a significant difference relative to the control (P < 0.05).
-
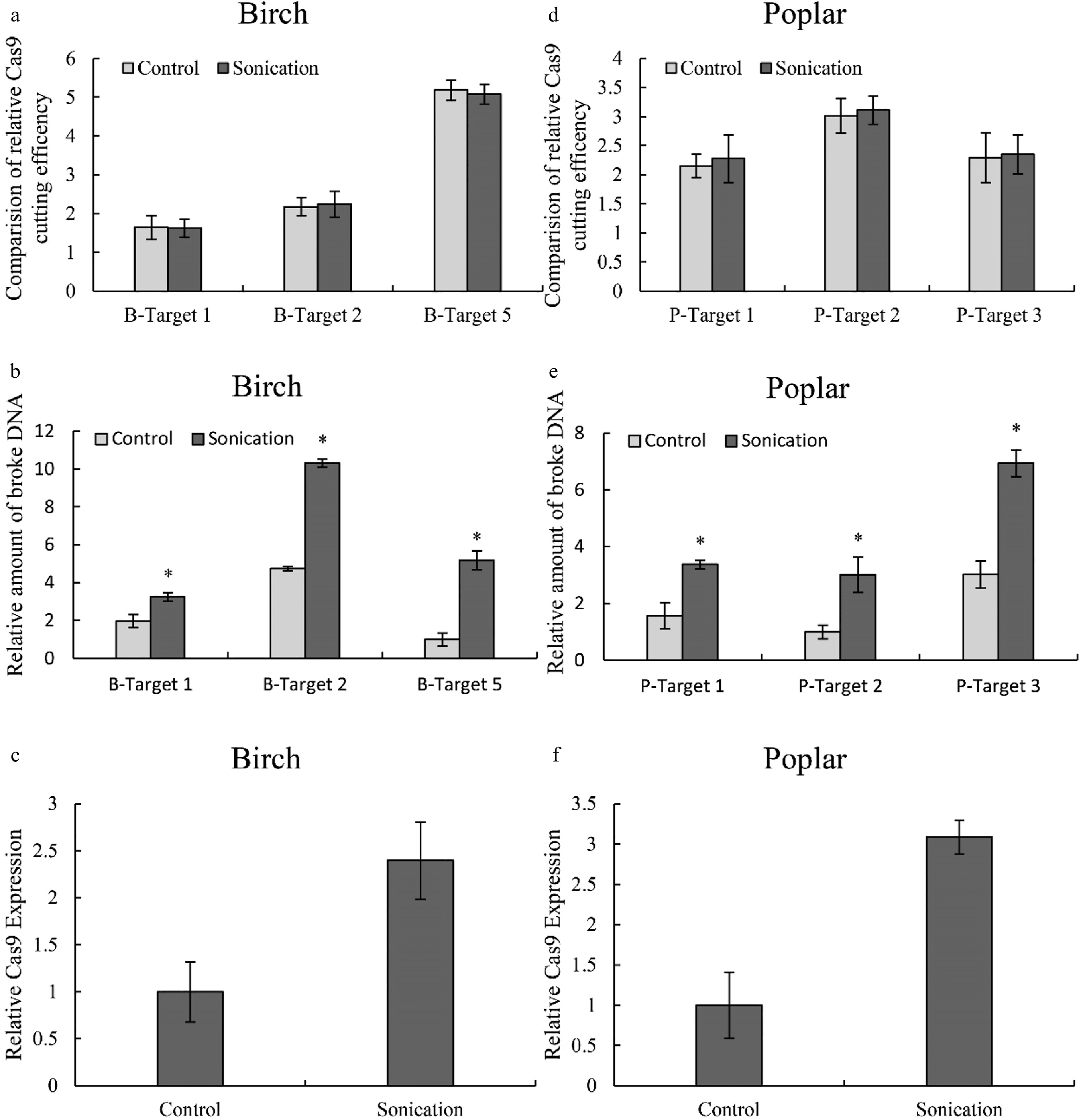
Figure 6. Determination of the effects of sonication on Cas9 cutting efficiency. (a, d): The effects of sonication on Cas9 cutting efficiency in birch (a) or poplar (d); (b, e): The relative amount of broken DNA in birch (b) or poplar (e) with or without sonication treatment. (c, f): Study of the expression of Cas9 in birch (c) or poplar (f) with or without sonication treatment. The plants were sonicated for 10 seconds, and the TCEP method was performed (Sonication). Plants without sonication were used as controls (Control). The number of target cutting sites of birch and poplar is consistent with Fig. 2 and Fig. 3, respectively. The target site with the lowest cutting efficiency or amount of broken DNA was set as 1 for normalization Cas9 cutting. Three target sites with high, medium and low CRISPR cutting efficiency were selected for study using qPCR. (c, f): The expression of the gene encoding Cas9 with or without sonication treatment. * indicates a significant difference compared with the control (p < 0.05).
Figures
(6)
Tables
(0)