-
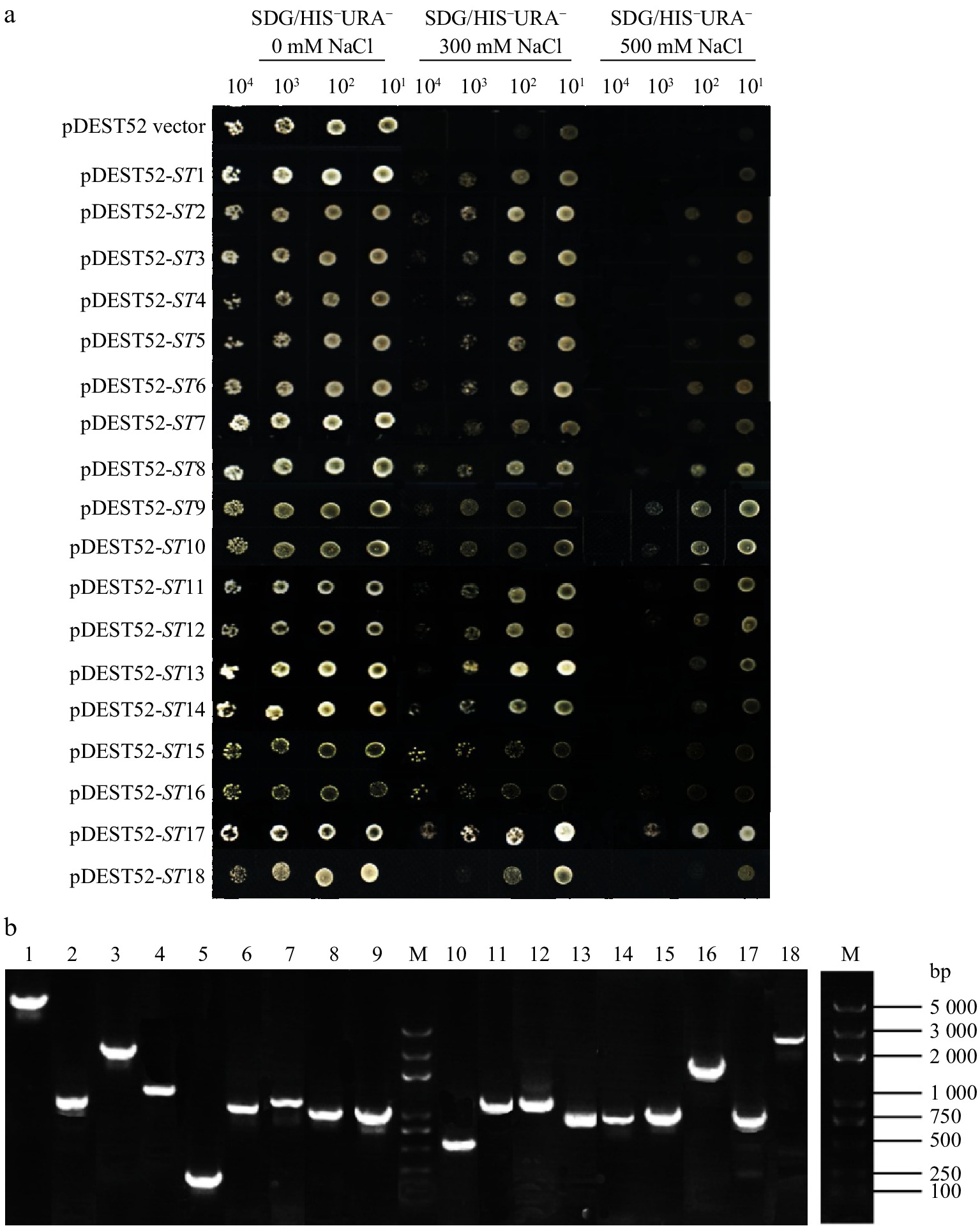
Figure 1. Salt-tolerance confirmation of 18 clones via library screening of alfalfa (a) and PCR detection of yeast plasmids (b).
-
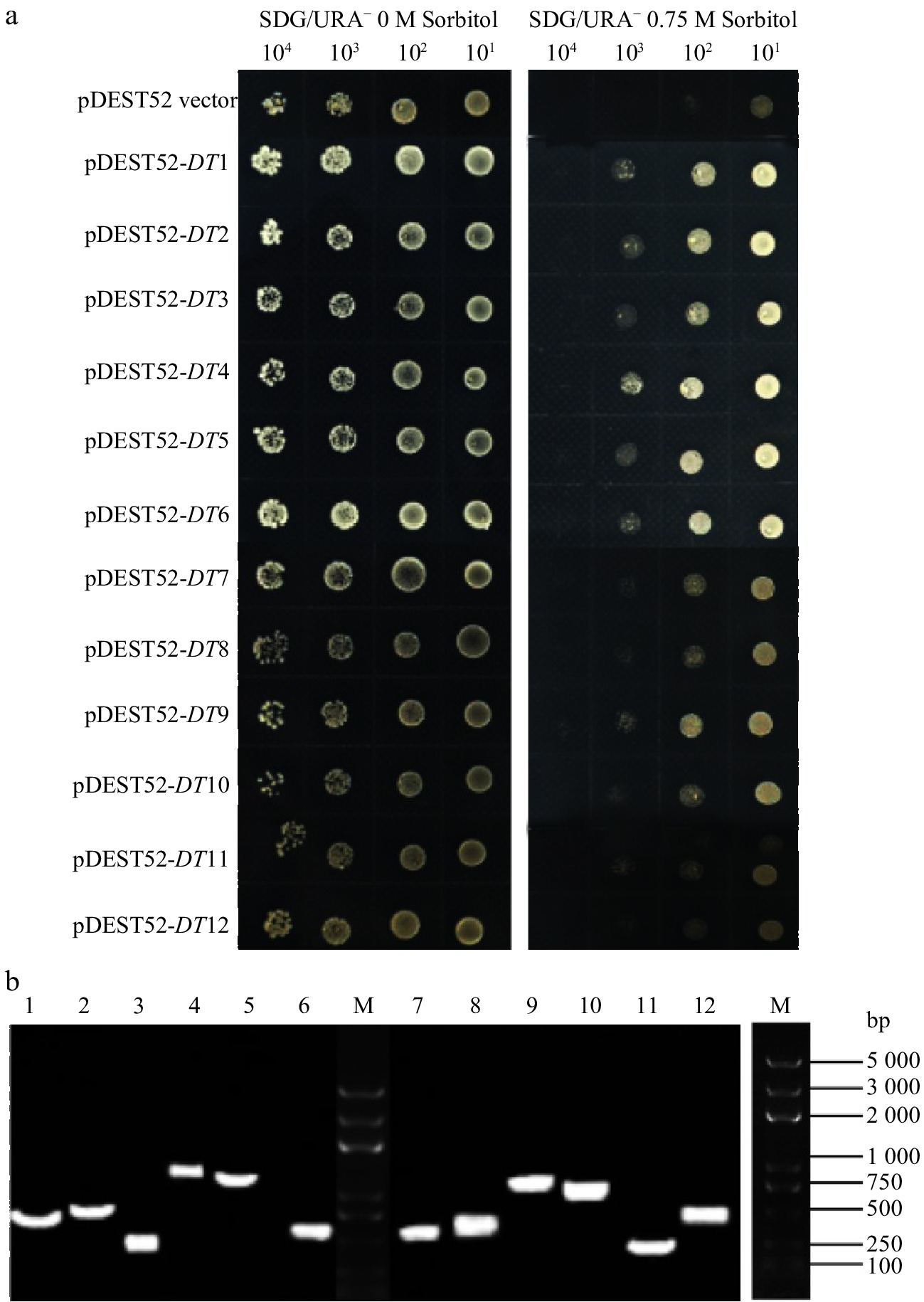
Figure 2. Drought-tolerance detection of 12 clones via library screening of alfalfa (a) and PCR analysis of yeast plasmids (b).
-
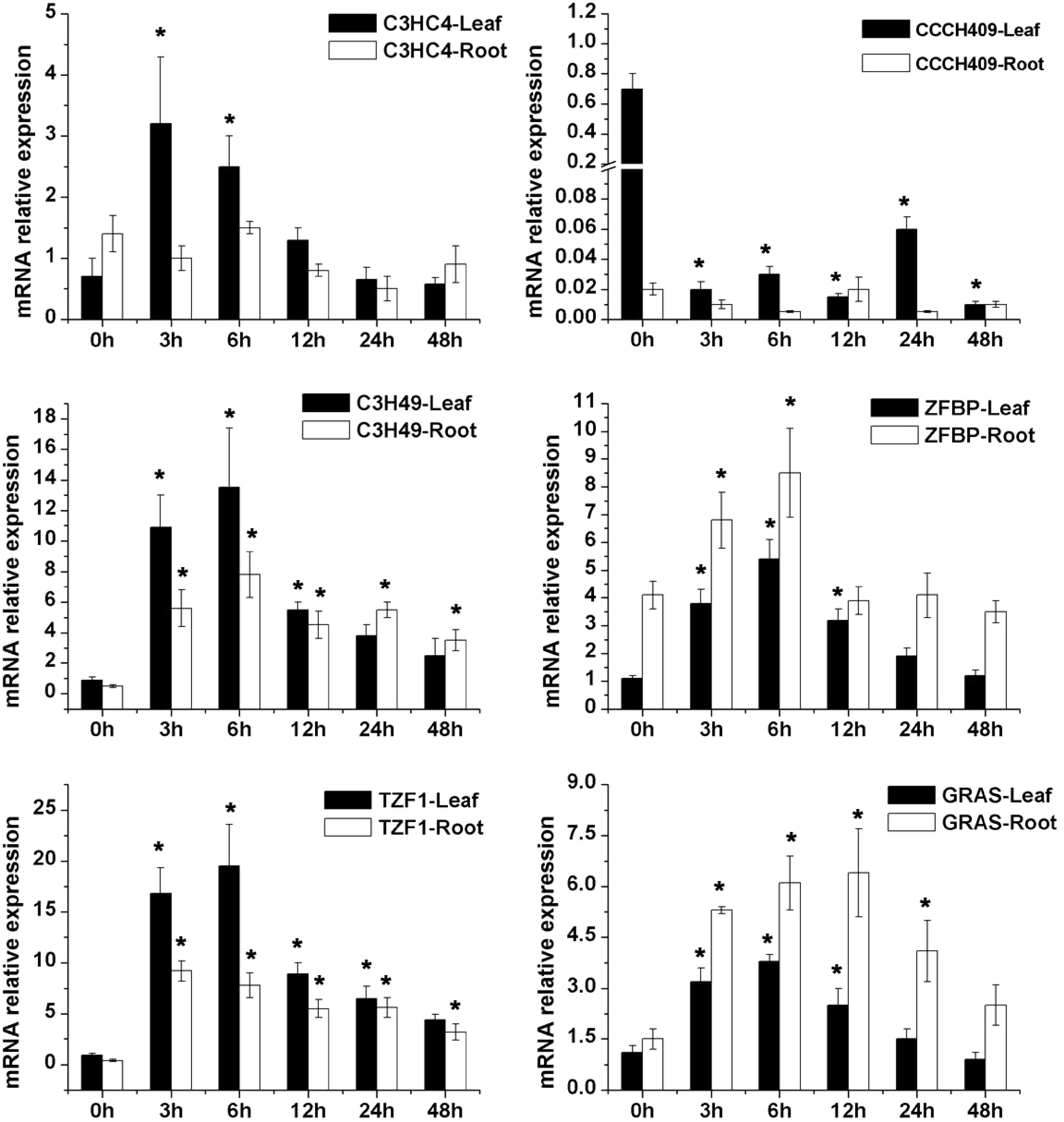
Figure 3. qRT-PCR analysis of six candidate salt-tolerant transcriptional factor of alfalfa including a GRAS family gene (GRAS) and five zinc finger protein (C3HC4, CCCH409, C3H49, ZFBP and TZF1) under 250 mM salt stress conditions. * Represent significant difference (p < 0.05) of salt stress samples for 3, 6, 12, 24 and 48 h compared to control sample (0 h), respectively.
-

Figure 4. qRT-PCR analysis of seven candidate salt-tolerant genes of alfalfa involved in protein phosphorylation modification (SLP, TAP46, PP1) and enzymes (LA, ACO1, bHFP and CYS1) under 250 mM salt stress conditions. * Represent significant difference (p < 0.05) of salt stress samples for 3, 6, 12, 24 and 48 h compared to control sample (0 h), respectively.
-

Figure 5. qRT-PCR analysis of five candidate salt-tolerant candidate genes of alfalfa participating in vesicle transporter (ERVT), transmembrane protein (LETM) and other genes (SCPT, ERD14, acpAP-2) under 250 mM salt stress conditions. * Represent significant difference (p < 0.05) of salt stress samples for 3, 6, 12, 24 and 48 h compared to control sample (0 h), respectively.
-
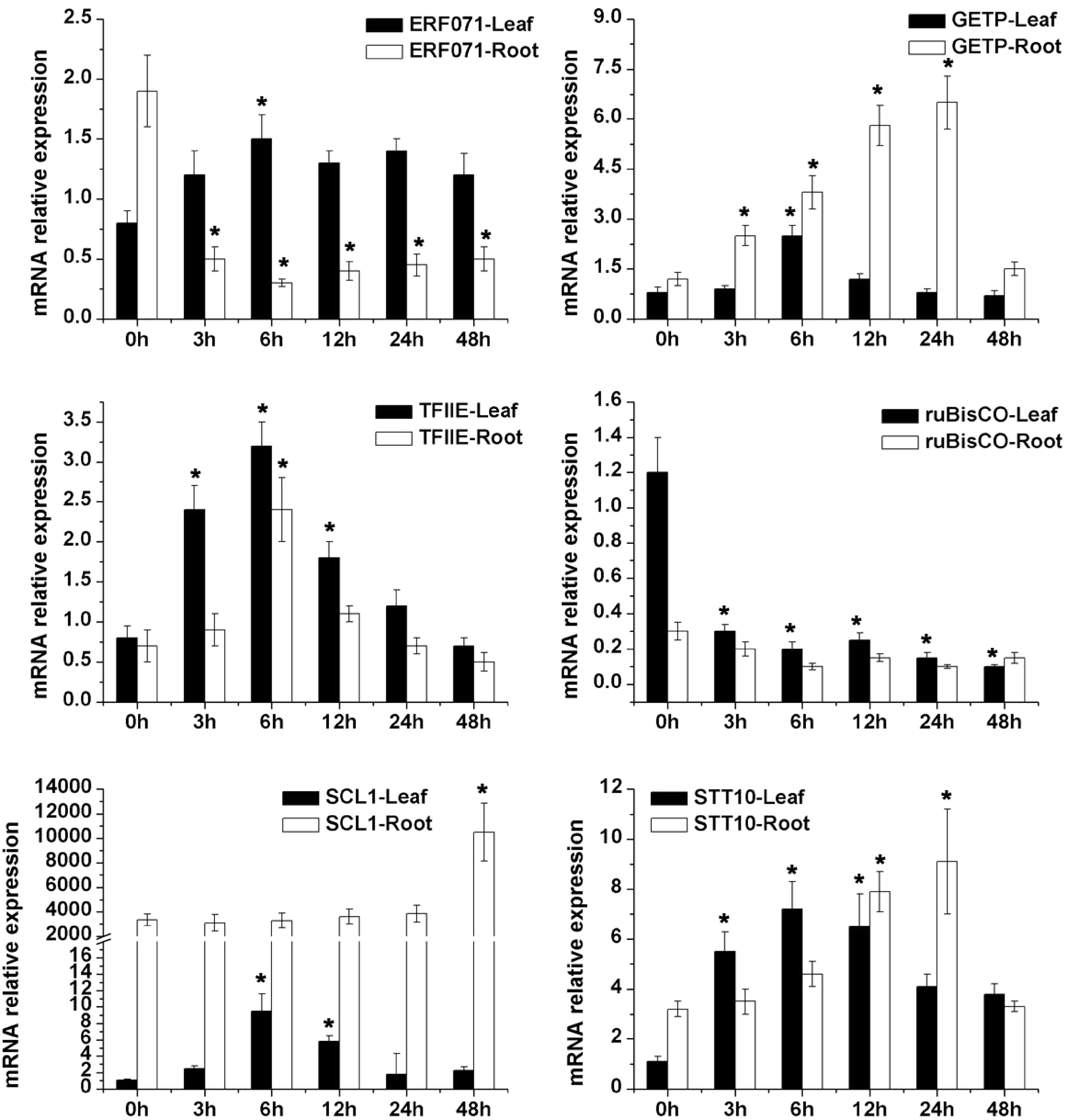
Figure 6. qRT-PCR analysis of six candidate drought-tolerant genes of alfalfa including transcriptional factor (ERF071, SCL, TFIIE, GETP), transport (STT10) and Rubisco under 20% PEG6000 simulated drought stress conditions. * Represent significant difference (p < 0.05) of drought stress samples for 3, 6, 12, 24 and 48 h compared to control sample (0 h), respectively.
-

Figure 7. qRT-PCR analysis of six candidate drought-tolerant genes of alfalfa including protein kinase (BSK7, CIPK11, cPGM), lipase (TGL1) and protease (cPDL1, POD10) under 20% PEG6000 simulated drought stress conditions. * Represent significant difference (p < 0.05) of drought stress samples for 3, 6, 12, 24 and 48 h compared to control sample (0 h), respectively.
-
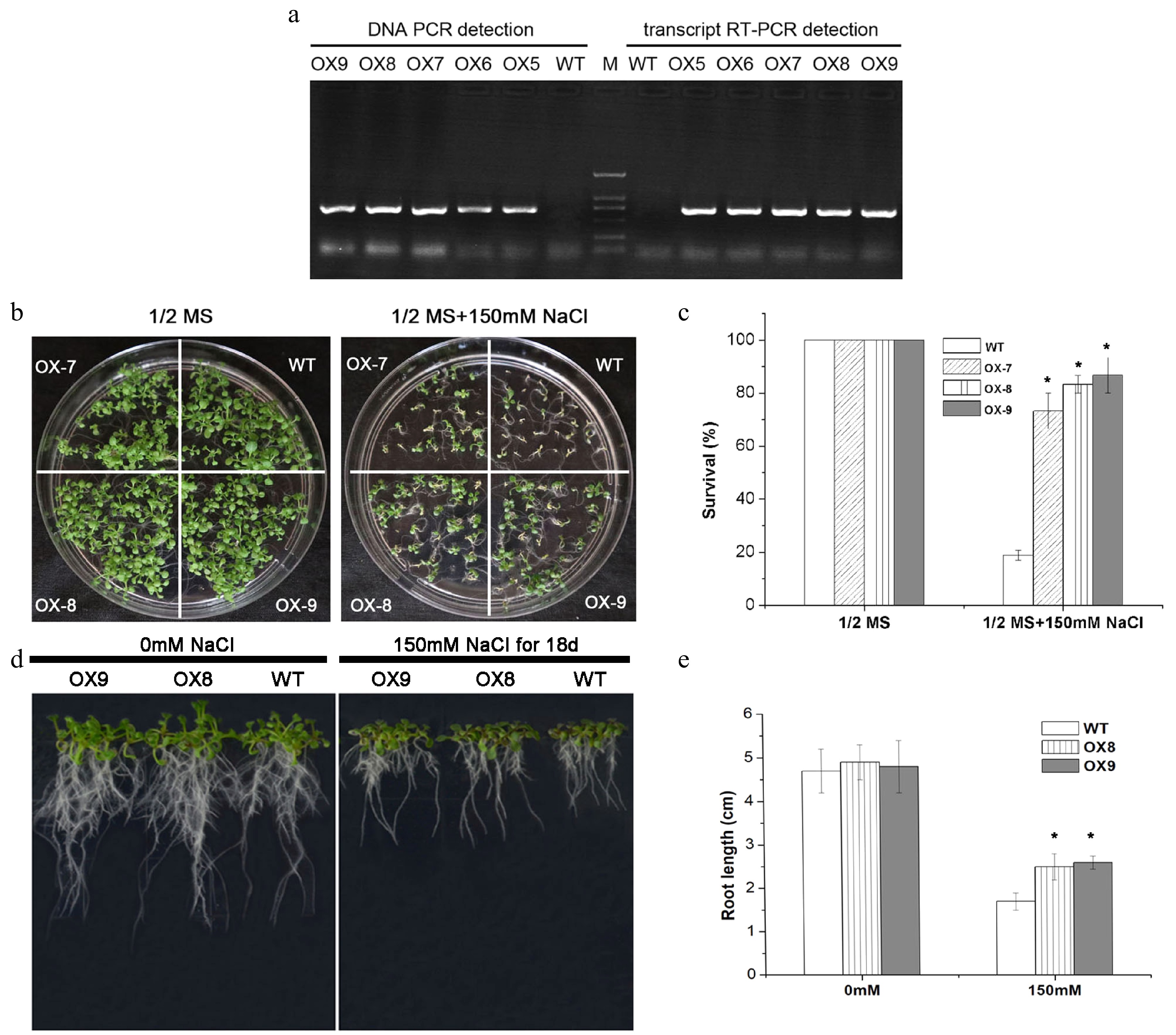
Figure 8. Salt-tolerance assays between wild type and ERVT transgenic lines of Arabidopsis. (a) Detection of PCR in DNA level and RT-PCR in mRNA level from wild type (WT) and transgenic plants (OX7, OX8, OX9). (b) Phenotype observation of plates. (c) Survival rate calculation under salinity stress with 0 and 150 mM NaCl. (d) Seedling growth in fritted clay under 0 and 150 mM NaCl treated conditions for 18 d. (e) Root length detection of seedlings. * Represent significant difference (p < 0.05) under salt stress compared to WT.
-

Figure 9. Drought-tolerance evaluation between wild type and CIPK11 transgenic lines of Arabidopsis. (a) Detection of PCR in DNA level and RT-PCR in mRNA level from wild type (WT) and transgenic plants (OX11, OX12, OX13). (b) Phenotype observation of plates. (c) Survival rate calculation under 0 and 200 mM mannitol simulated drought stress. (d) Seedling growth in fritted clay under normal watering and drought treated conditions for 14 d. (e) Root length detection of seedlings. * Represent significant difference (p < 0.05) under drought stress compared to WT.
-
Salt tolerance clones Amino acid (aa) Protein function prediction GenBank accession number ST1 3167 Spatacsin carboxy-terminus protein (SCPT) MZ760872 ST2 358 L-allo-threonine aldolase-like protein (LA) MZ760873 ST3 1026 Adaptor protein complex AP-2, alpha subunit (acpAP-2) MZ760874 ST4 592 GRAS family transcription factor (GRAS) MZ760875 ST5 98 Cystatin (CYS1) MZ760876 ST6 353 Endoplasmic reticulum vesicle transporter protein (ERVT) MZ760877 ST7 409 Zinc finger CCCH domain protein (CCCH409) MZ760878 ST8 325 Alpha/beta hydrolase family protein (bHFP) MZ760879 ST9 325 Zinc finger, C3HC4 type (RING finger) protein (C3HC4) MZ760880 ST10 222 EARLY RESPONSE TO DEHYDRATION 14 (ERD14) MZ760881 ST11 403 PP2A regulatory subunit TAP46-like protein (TAP46) MZ760882 ST12 469 SCP1-LIKE SMALL PHOSPHATASE 4, SSP4 (SLP) MZ760883 ST13 299 Zinc finger B-box protein (ZFBP) MZ760884 ST14 379 Zinc finger CCCH domain protein (TZF1) MZ760885 ST15 326 Serine/threonine-protein phosphatase PP1 (PP1) MZ760886 ST16 724 Leucine zipper EF-hand transmembrane protein (LETM) MZ760887 ST17 377 Zinc finger CCCH domain protein (C3H49) MZ760888 ST18 901 Cytoplasmic-like aconitate hydratase (ACO1) MZ760889 Table 1. Sequence analysis and function prediction of 18 salt tolerant candidate genes screened from cDNA expression library of alfalfa.
-
Drought tolerance clones Amino acid (aa) Protein function prediction GenBank accession number DT1 422 CBL-interacting serine/threonine-protein kinase 11 (CIPK11) MZ760890 DT2 592 Scarecrow-like protein 1 (SCL1) MZ760891 DT3 326 Peroxidase 10 (POD10) MZ760892 DT4 259 Ethylene-responsive transcription factor ERF071 (ERF071) MZ760893 DT5 704 Triacylglycerol lipase 1 (TGL1) MZ760894 DT6 447 Elongation factor 1-alpha (GETP) MZ760895 DT7 265 Transport and Golgi organization 2 homolog (STT10) MZ760896 DT8 333 Cytoplasmic phosphoglucomutase (cPGM) MZ760897 DT9 274 Brassinosteroid-signaling kinase BSK7 (BSK7) MZ760898 DT10 281 General transcription factor IIE subunit 2 (TFIIE) MZ760899 DT11 350 Rubisco large subunit-binding protein subunit alpha, chloroplastic (Rubisco) MZ760900 DT12 432 Protease Do-like 1, chloroplastic (cPDL1) MZ760901 Table 2. Sequence analysis and function prediction of 12 drought tolerant candidate genes screened from cDNA expression library of alfalfa.
Figures
(9)
Tables
(2)