-

Figure 1.
The pan-plastome of Nelumbo. The inner genes of the outer circle are transcribed counterclockwise while the outer genes are transcribed clockwise; genes with introns were marked with an asterisk (ycf3, clpP, and rps12 contain two introns all others contain one). The GC content is displayed as gray bars inside of the tetrad divisions (LSC, SSC, IRA, and IRB) SNVs, InDels, and block substitutions are represented within the GC content as orange, blue, and yellow lines, respectively. Comparisons of the complete genome length and each of the four subregions (given in median lengths) are compared for each of the different Nelumbo types N. lutea LA, LW, LF, LS, and LR.
-

Figure 2.
Variant locations categorized by genic position (CDS, Introns, and IGS) and functional group.
-
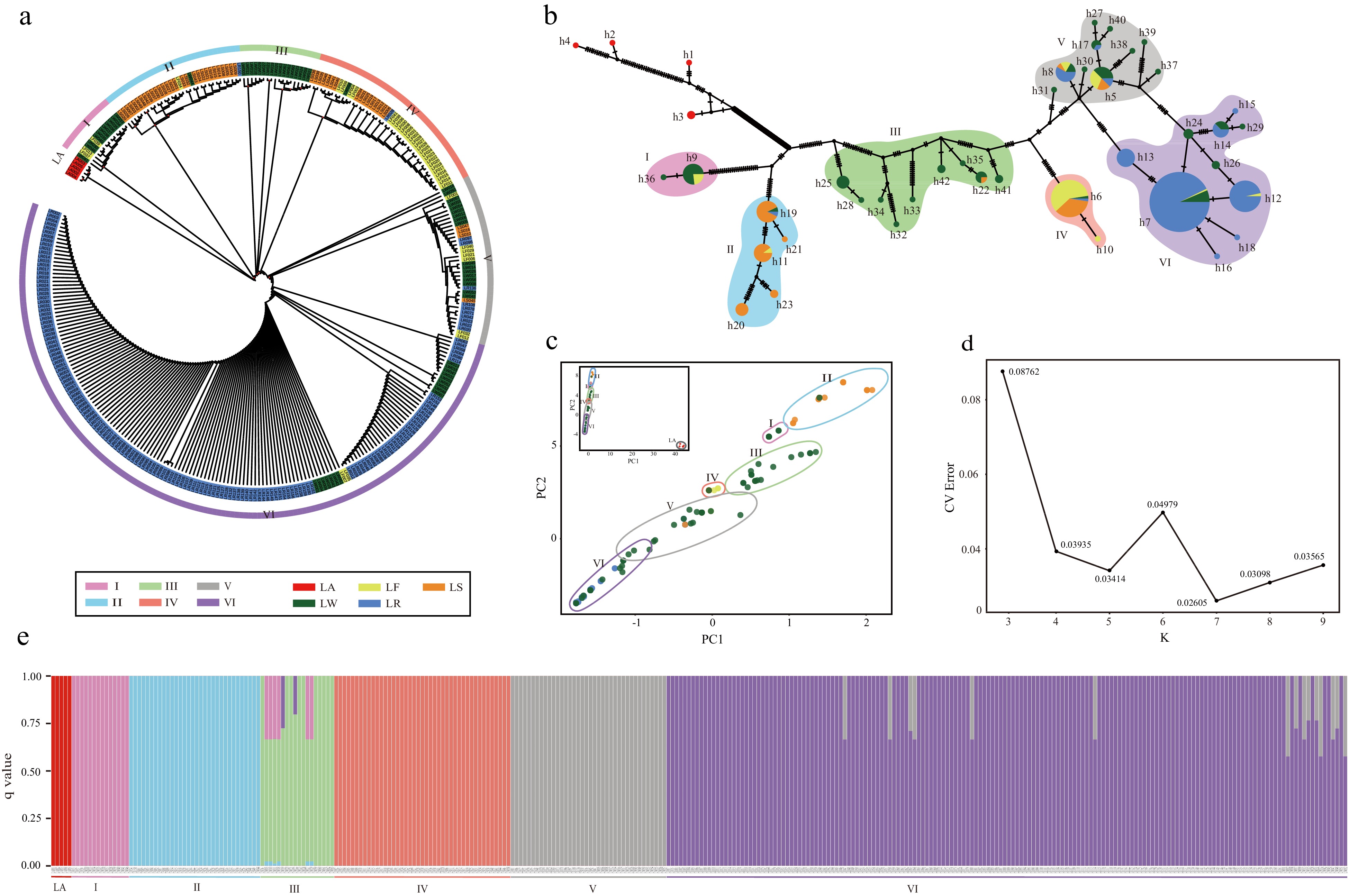
Figure 3.
Population structure of Nelumbo. All analyses resolved similar membership into six genetic clusters (I−VI). (a) ML tree of 316 accessions. (b) Median-joining network with haplotype identifiers adjacent to nodes, size of pie chart proportional to number of accessions sharing the same haplotype, colors in the pie chart represent percentage of accessions from a given Asian lotus type. (c) PCA analysis including LA (upper left inset) and excluding LA showing the first two components in both cases. (d) CV errors across a range of K values from 3−9. (e) Population structure bar-plot at K = 7.
-
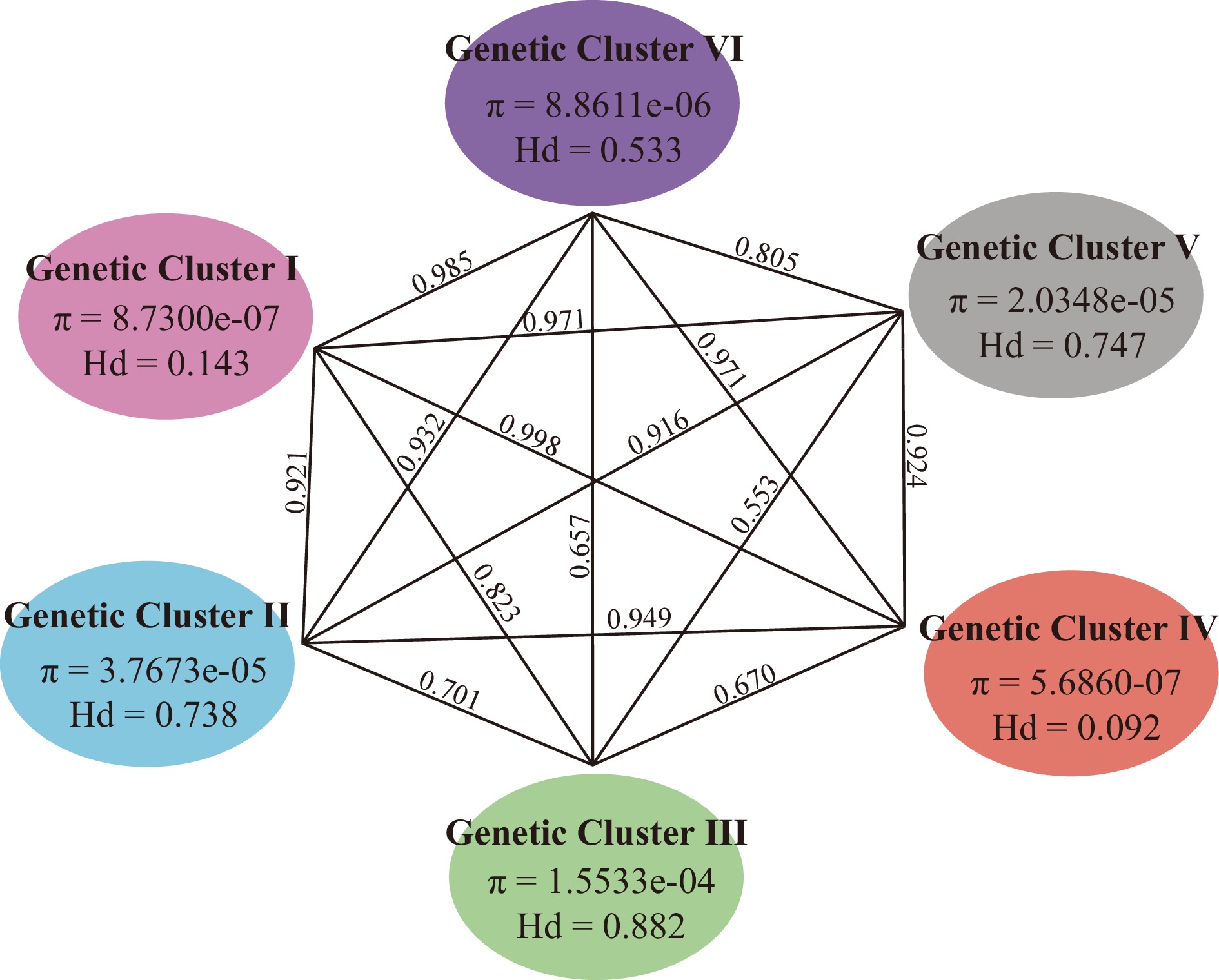
Figure 4.
Genetic diversity and differentiation of six genetic clusters of Asian lotus. Numbers above lines connecting two bubbles represent pairwise Fst calculated between respective genetic clusters.
-
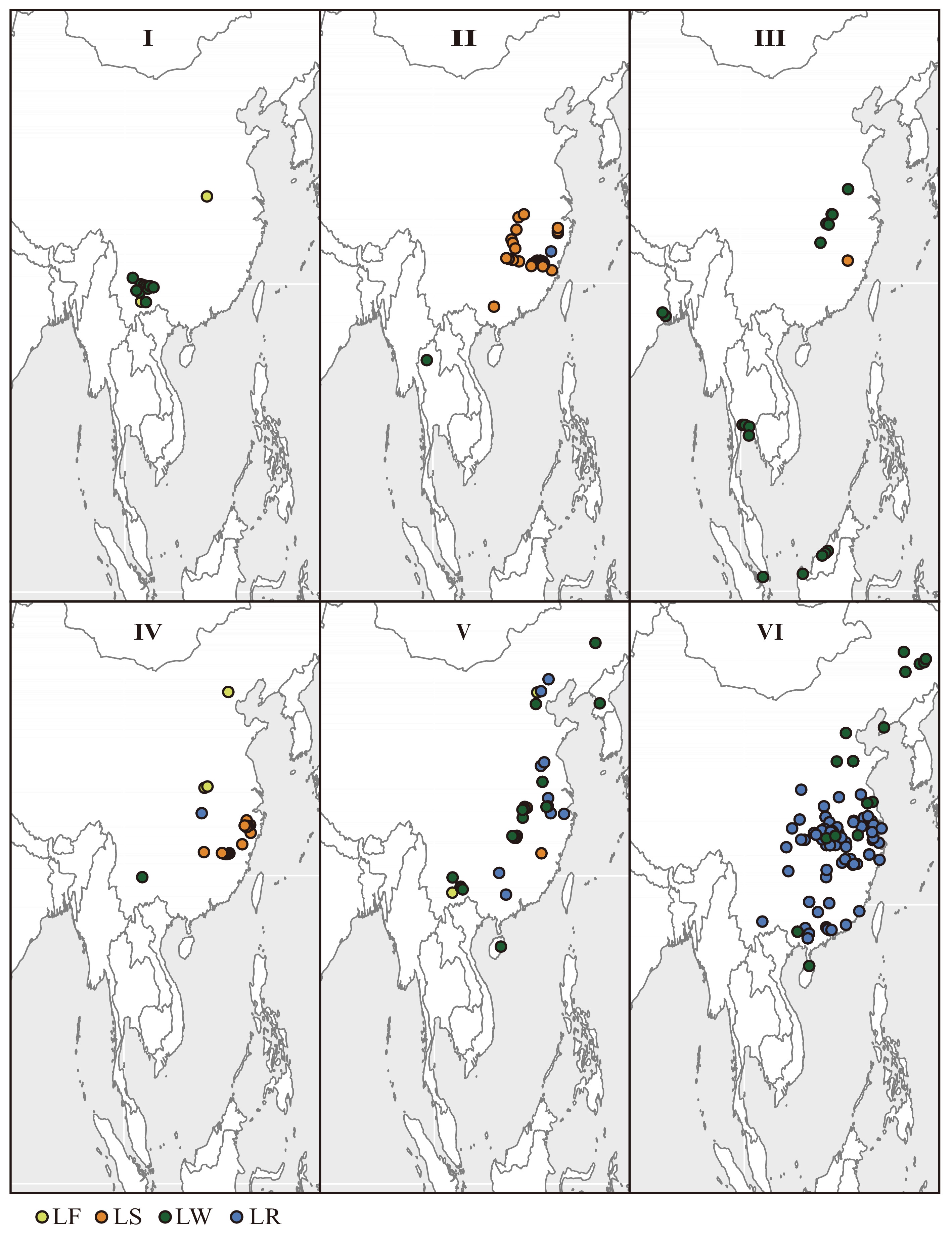
Figure 5.
Geographic distribution of Asian lotus collections used in this study separated by genetic cluster and color coded by type (see Fig. 3).
-
Variants Total Region Location LSC SSC IRA/B CDS Intron IGS SNV 418 (294) 274 (182) 112 (89) 16 (12) 117 (91) 33 (23) 268 (180) Block substitution 70 (54) 56 (42) 12 (11) 1 (0) 3 (3) 4 (4) 63 (46) InDel 208 (151) 157 (118) 43 (27) 4 (3) 4 (2) 25 (19) 179 (130) Total 696 (499) 487 (342) 167 (127) 21 (15) 124 (96) 62 (46) 510 (356) Table 1.
Number of variants among 316 Nelumbo accessions. The number of variants found only in N. nucifera are indicated in parenthesis.
Figures
(5)
Tables
(1)