-
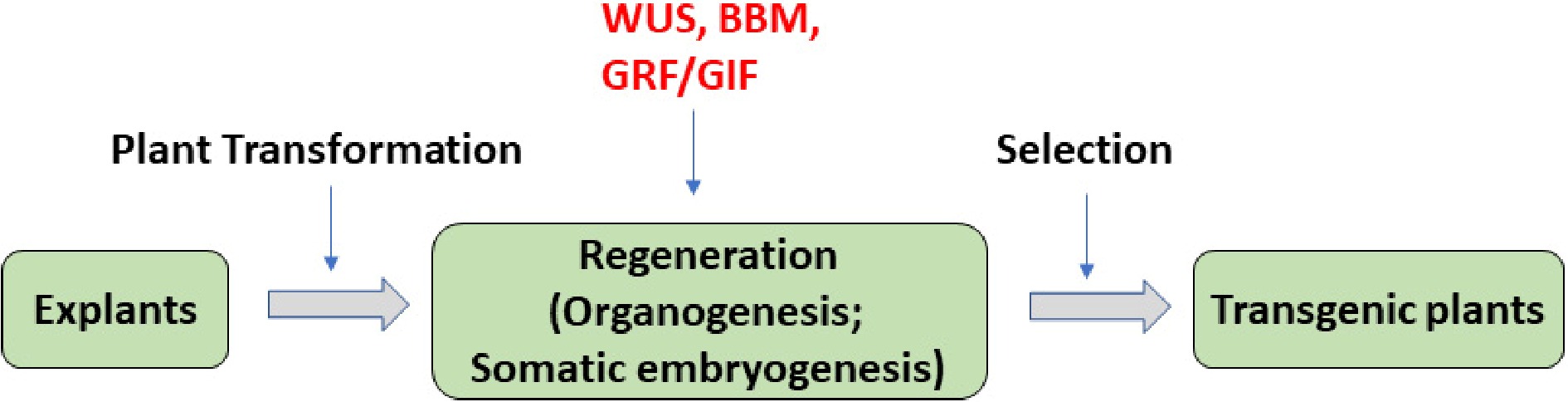
Figure 1.
The regeneration-promoting effect of WUS, BBM, GRFs, and GRFs–GRFs genes in plant transformation. In standard plant transformation systems, transgene is delivered into selected explants, plant tissue culture is used to induce plant regeneration, then the final transgenic plants are selected from regenerated plants. Among them, plant regeneration is often the bottleneck of the process. Genes WUS, BBM, GRFs, and GRFs–GRFs (red) could promote plant regeneration via either organogenesis or somatic embryogenesis in various plant species.
-
Gene* Promoter Explants Effects Ref. AtWUS Estrogen-inducible A. thaliana root High somatic embryo formation frequency [15] Estrogen-inducible Nicotiana tabacum leaf Shoot formation from root tip [20] 35S Gossypium hirsutum hypocotyl Shoot formation from root tip [16] vsp1 Medicago truncatula seedling radicle 47.75% increase in embryogenic callus formation [18] ZmWUS2 ZmPLTP Zea mays immature embryo Enhanced callogenesis and embryogenesis [66] Nos A. thaliana (seedling), Solanum lycopersicum (seedling), N. tabacum (seedling/mature plant), Solanum tuberosum (mature plant), Vitis. vinifera (mature plant) de novo meristem induction [38] AtWUS-GR, AtSTM-GR 35S A. thaliana (floral dip) Triggered ectopic organogenesis [18] AtWUS, CHAP3A (PmLEC1) Estrogen-inducible Picea glauca immature embryo Did not induce somatic embryogenesis [59] eGFP-GhWUS1a, eGFP-GhWUS1b Estrogen-inducible G. hirsutum hypocotyl Inhibited embryogenic callus formation [60] AtBBM, BnBBM 35S, inducible N. tabacum leaf Enhance the regeneration capacity [24] BcBBM 35S Populus tomentosa calli Plant regeneration through somatic embryogenesis [25] BnBBM 35S, HnUbB1 A. thaliana (floral dip) B. napus haploid embryo Spontaneous formation of somatic embryos and cotyledon-like structures [22] BnBBM
EgAP2-1 (BBM)35S Capsicum. annuum cotyledon Made recalcitrant pepper transformable [23] 35S A. thaliana (floral dip) Enhanced regeneration capacity [63] GmBBM1 35S A. thaliana (floral dip) Induced somatic embryos on vegetative organs [64] TcBBM 35S A. thaliana (floral dip) Enhanced/hormone-independent somatic [65] AtBBM-GR 35S A. thaliana (floral dip) Improved plant regeneration for extended periods of time in tissue culture [62] HvWUS, HvBBM ZmAxig1, ZmPLPT Hordeum vulgare Co-expression increased transformation efficiency by 3 times [61] ZmBBM+ZmWUS2 ZmUbi, Nos Z. mays immature embryo, mature embryo, seedling leaf segment; Oryza sativa calli; Sorghum bicolor immature embryo; Saccharum officianrum calli Enabled transformation of recalcitrant varieties and/or increased transformation efficiency [26−28] ZmAxig1, ZmPLTP Z. mays immature embryo Established rapid callus-free transformation [29] ZmPLTP S. bicolor immature embryo Reduced genotype dependence, accelerated regeneration, increased transformation efficiency [67] AtGRF5/BvGRF5-L 2×35S Beta. vulgaris cotyledon, hypocotyl Enabled transformation of recalcitrant varieties. Increased transformation efficiency [33] AtGRF5/HaGRF5-L 2×35S Helianthus annuus cotyledon Improved transgenic shoot formation GmGRF5-L PcUbi4-2 Glycine. max primary node Improved transgenic shoot formation BnGRM5-L PcUbi4-2 B. napus hypocotyl Promoted callus production ZmGRF5-L1/2 BdEF1 Z. mays immature embryo) Increased transformation efficiency ~3 times TaGRF4-GIF1 ZmUbi Triticum aestivum immature embryo Increased regeneration efficiency 7.8 times; shortened protocol [34] O. sativa calli from seeds Increased regeneration efficiency 2.1 times ClGRF41-GIF1/VvGRF4-GIF1 35S Citrus limon etiolated epicotyl Increased regeneration efficiency ~4.7 times CIGRF42-GIF1 35S Citrullus lanatus cotyledon Increased transformation efficiency ~9 times [68] *At, A. thaliana; Zm, Z. mays; Pm, Picea mariana; Gh, G. hirsutum; Bn, B. napus; Bc, B. campestris; Eg, Elaeis guineensis; Gm, G. max; Tc, Theobroma cacao; Hv, H. vulgare; Bv, B. vulgaris; Ta, T. aestivum; Cl, 1C. limon, 2C. lanatus; Vv, V. vinifera. Table 1.
The effects of WUS, BBM, GRFs, and GRFs–GRFs on plant development and genetic transformation.
Figures
(1)
Tables
(1)