-
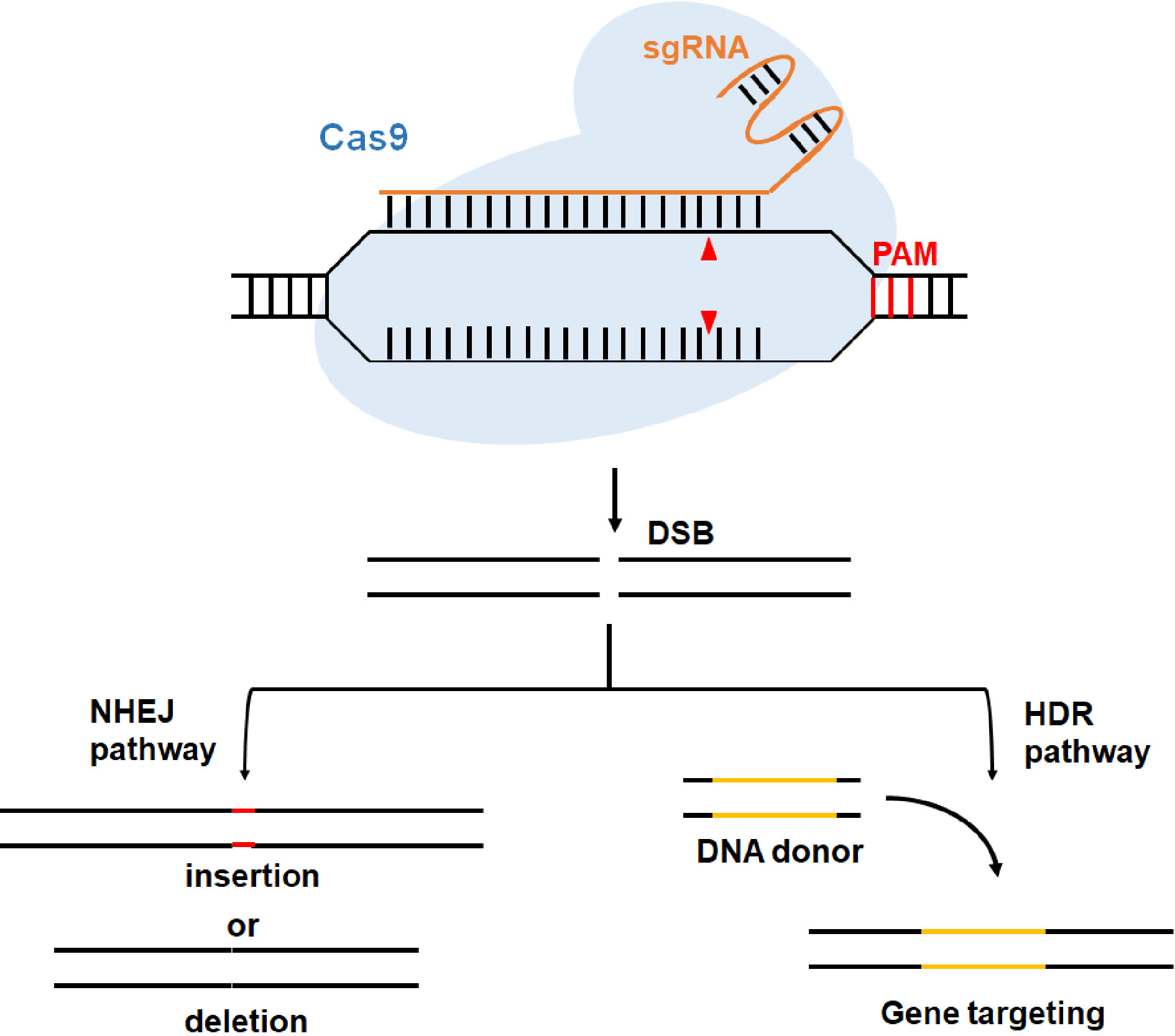
Figure 1. Schematic diagram of genome editing induced by CRISPR/Cas9 system. Cas9 protein cleaves the target sequence (the cut sites are indicated by red triangles) that is complementary with the single guide RNA (sgRNA) and produces double strand breaks (DSB). The DSB could be repaired through non-homologous end joining (NHEJ) or the homology-directed repair (HDR) pathway, resulting in indel (insertion or deletion) or gene targeting mutation.
-

Figure 2. Schematic overview of base editing and prime editing. In the cytosine base editor (CBE) system, a Cas9 nickase (Cas9n) is fused with a cytidine deaminase and a uracil DNA glycosylase inhibitor (UGI). The CBE could mediate C-to-T substitutions. In the adenine base editor (ABE) system, a Cas9n protein is fused with an engineered Escherichia coli adenosine deaminase (ecTadA), which catalyzes the conversion of adenine (A) to inosine (I). The I is recognized as guanine (G) by DNA polymerase during replication, resulting in A-to-G substitutions. Prime editing (PE) is accomplished by M-MLV-Cas9n-pegRNA complex, in which pegRNA functions as a guide RNA and also provides primer binding site (PBS) and reverse transcriptase (RT) template. The nicked target DNA sequence hybridizes to the PBS, priming reverse transcription of the template into the DNA sequence. The desired mutations within the pegRNA are indicated in purple. PAM, protospacer adjacent motif.
-
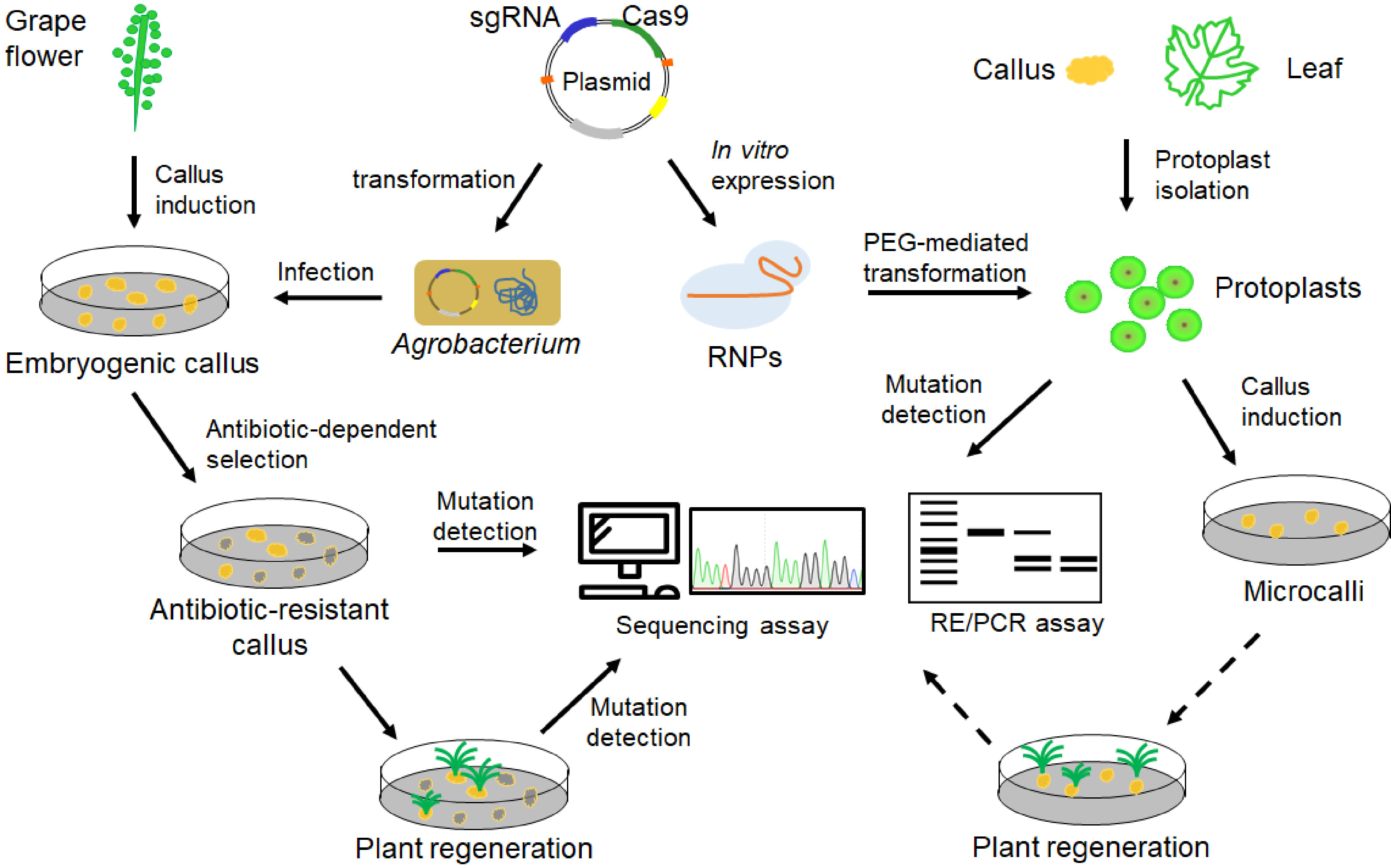
Figure 3. Pipeline of CRISPR/Cas9-mediated genome editing in grape. Both embryogenic callus and protoplasts could be used as materials for grape genome editing. CRISPR/Cas9 reagents such as plasmid constructs and CRISPR/Cas9 ribonucleoproteins (RNPs) can be delivered into the embryogenic callus and protoplasts, respectively. Edited grapevine plants could regenerate from embryogenic callus. Microcalli could be induced from grape protoplasts, but plant regeneration from the protoplasts-induced calli (indicated by broken lines) has not been achieved.
-
Target
genessgRNA/Cas9 promoter Explant Delivery method Modification Target traits Editing efficiency Off-target effect Reference IdnDH AtU6/35S Embryogenic cells of 'Chardonnay' Agrobacterium-mediated transformation KO Tartaric acid synthesis 100% No Ren et al.[12] MLO-7 Not mentioned Embryogenic calli of 'Chardonnay' PEG-mediated transformation KO Powdery mildew resistance 0.1% ND Malnoy
et al.[13]VvPDS AtU6/
PcUbi4-2Embryonic calli of
'Neo Muscat'Agrobacterium-mediated transformation KO Albino phenotype 2%−86% No Nakajima et al.[81] VvPDS AtU6/2×35S Embryogenic cells of 'Chardonnay' and
'41B'Agrobacterium-mediated transformation KO Albino phenotype 22.2%−59.9% ('Chardonnay')
30.3%−86.6% ('41B')ND Ren et al.[89] VvWRKY52 AtU3, AtU6/2×35S Proembryonal masses (PEM) of 'Thompson Seedless' Agrobacterium-mediated transformation KO Botrytis cinerea resistance 31% No Wang
et al.[83]CCD8 AtU6/35S Embryogenic cells
of '41B'Agrobacterium-mediated transformation KO Strigolactones biosynthesis and shoot branching 66.7% No Ren et al.[82] VvPR4b AtU6/35S PEM of 'Thompson Seedless' Agrobacterium-mediated transformation KO Powdery mildew resistance 68.8% No Li et al.[85] VvMLO3, VvMLO4 AtU3, AtU6/2×35S PEM and somatic embryos of 'Thompson Seedless' Agrobacterium-mediated transformation KO Powdery mildew resistance 12.8%−38.5% ND Wan
et al.[84]TAS4,
MYBA7MtU6.6/
ZmUbiEmbryogenic callus
of rootstock '101-14'Agrobacterium-mediated transformation KO Anthocyanin accumulation related to Pierce disease (PD) and Grapevine Red Blotch Virus (GRBV) Not mentioned Yes Sunitha & Rock[80] PDS
TMT1,
TMT2AtU6, VvU3, VvU6/35S, VvUbi Embryogenic cells of '41B' Agrobacterium-mediated transformation KO Albino phenotype
Sugar accumulation23.5%−43.2% (PDS)
10.4%−20.9% (TMTs)ND Ren et al.[90] Abbreviations: AtU6, Arabidopsis thaliana U6 promoter; MtU6.6, Medicago truncatula U6.6 promoter; VvU6, Vitis vinifera U6 promoter; PcUbi4-2, Petroselinum crispum ubiquitin4-2 promoter; ZmUbi, Zea mays ubiquitin promoter; VvUbi, V. vinifera ubiquitin promoter; 35S, CaMV 35S promoter; KO, Knock out; ND, Not determined. Table 1. Applications of the CRISPR/Cas9 system in grape.
Figures
(3)
Tables
(1)