-
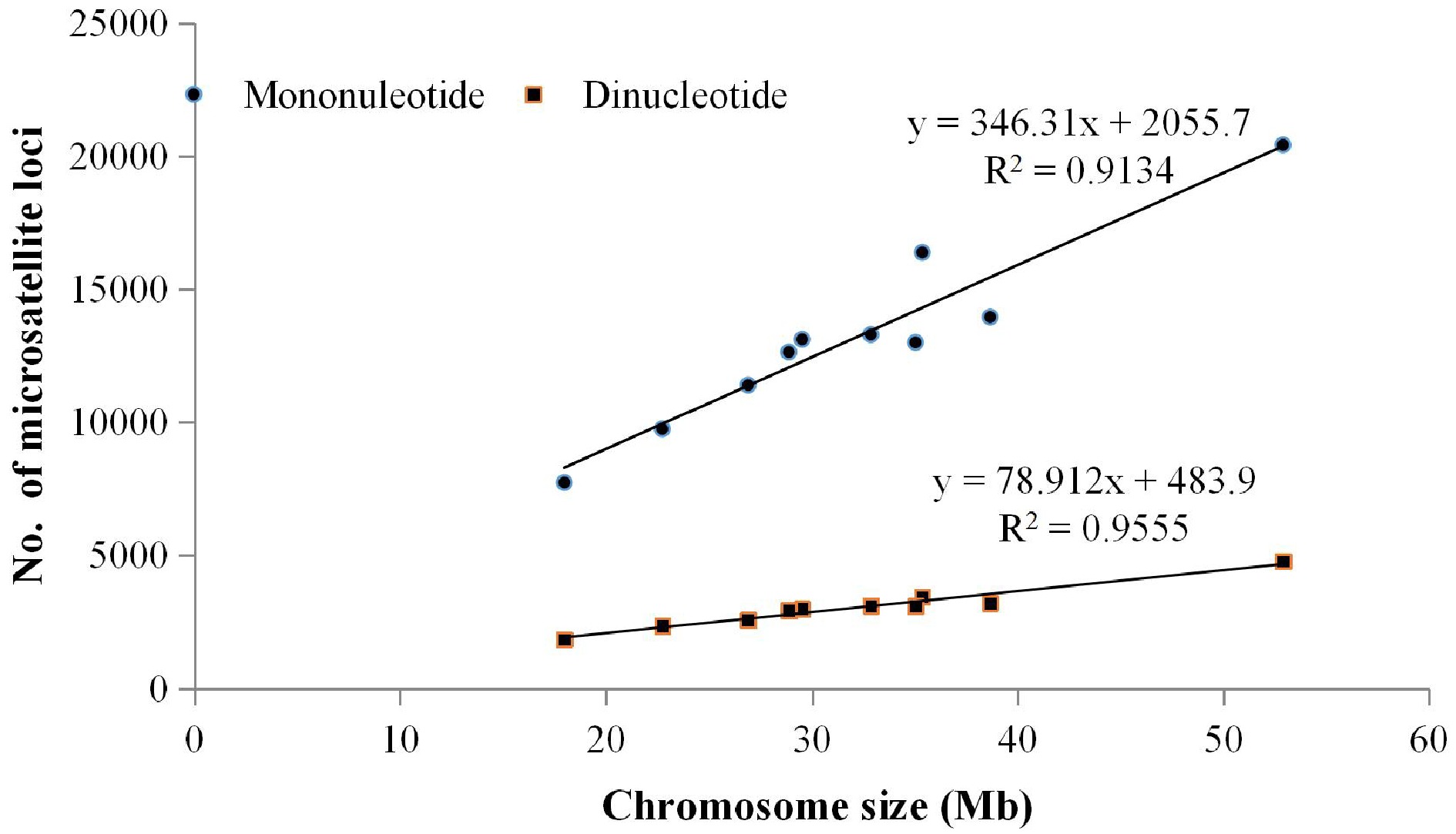
Figure 1.
Correlation analysis between chromosome size and abundance of the mono-nucleotide and di-nucleotide repeat motifs.
-
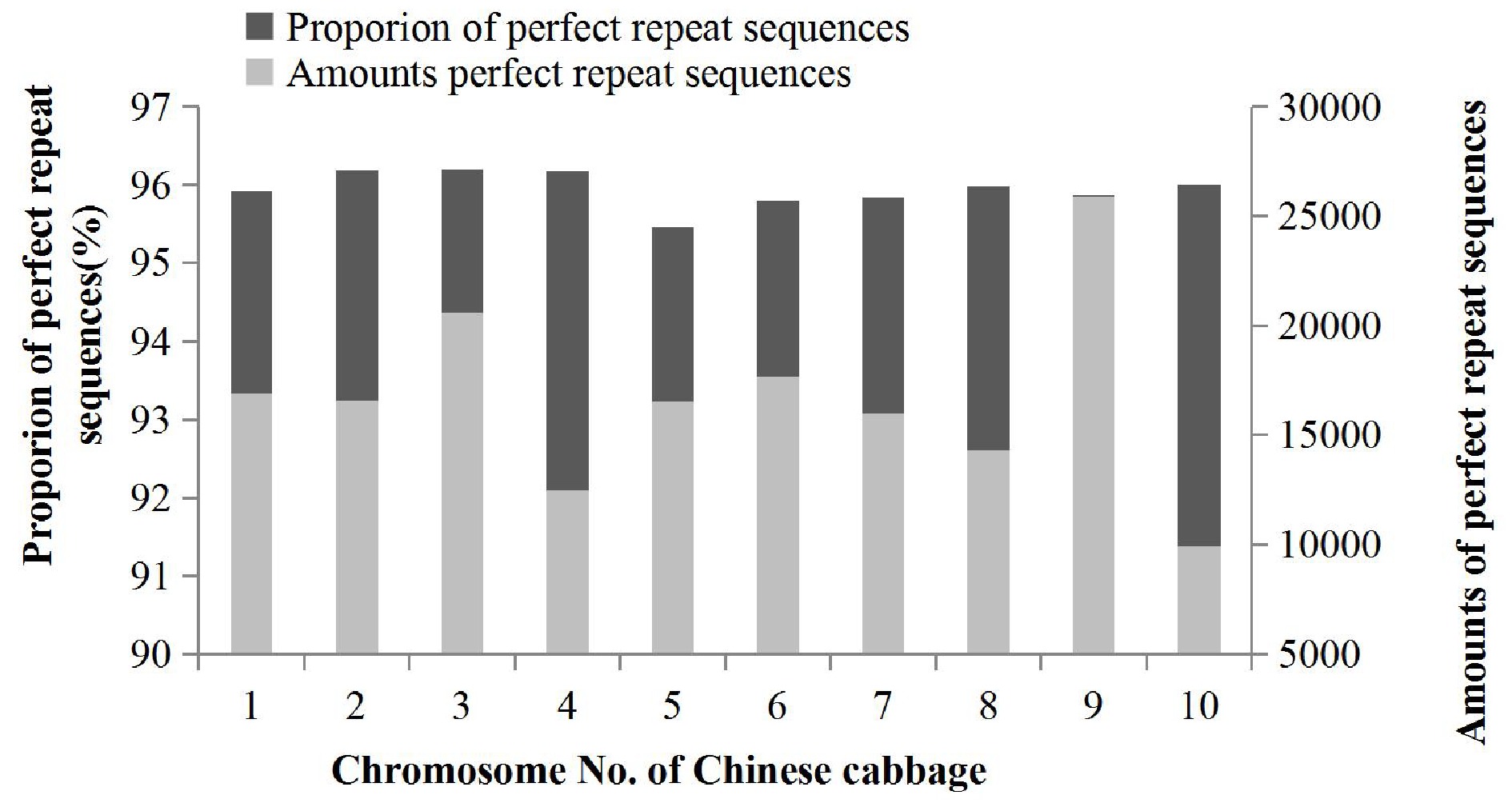
Figure 2.
Distribution of perfect repeat sequences in the Chinese cabbage genome.
-
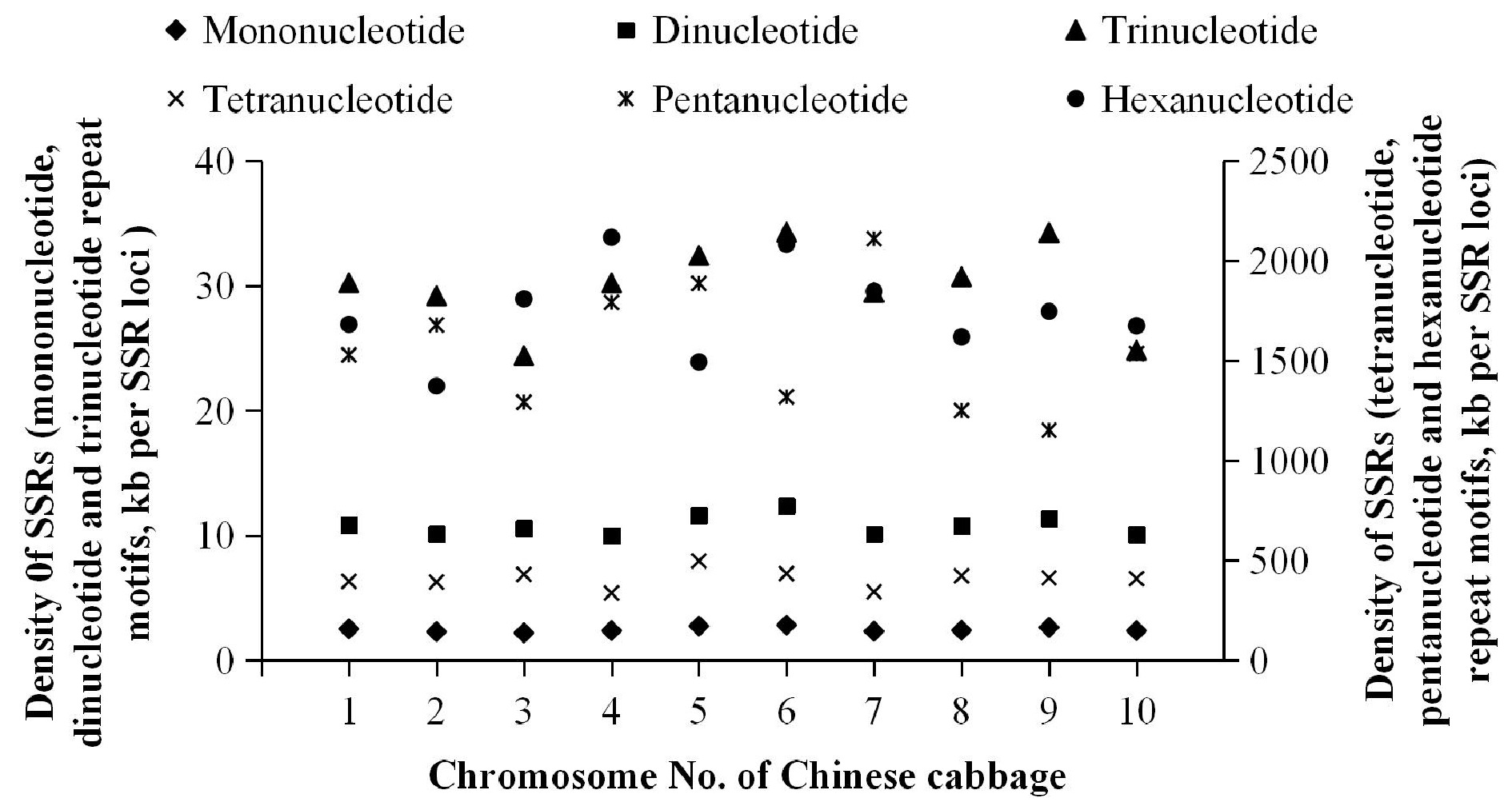
Figure 3.
Density of SSRs in Chinese cabbage.
-
Chromosome no. Chromosome size (Mb) Repeat type Total Mono-
nucleotideDi-
nucleotideTri-
nucleotideTetra-
nucleotidePenta-
nucleotideHexa-
nucleotide1 32.85 13,291/404.60 3,092/94.12 1,113/33.88 85/2.59 22/0.67 20/0.61 17,623/536.47 2 29.51 13,111/444.29 2,974/100.78 1,035/35.07 77/2.61 18/0.61 22/0.75 17,237/584.11 3 35.35 16,379/463.34 3,424/96.86 1,484/41.98 84/2.38 28/0.79 20/0.57 21,419/605.91 4 22.75 9,743/428.26 2,330/102.42 771/33.89 69/3.03 13/0.57 11/0.48 12,937/568.66 5 35.02 12,994/371.05 3,083/88.04 1,106/31.58 72/2.06 19/0.54 24/0.69 17,298/493.95 6 38.65 13,949/360.91 3,192/82.59 1,154/29.86 91/2.35 30/0.78 19/0.49 18,435/476.97 7 28.86 12,633/437.73 2,914/100.97 1,003/34.75 86/2.98 14/0.49 16/0.55 16,666/577.48 8 26.88 11,388/423.66 2,549/94.83 896/ 33.33 65/2.42 22/0.82 17/0.63 14,937/555.69 9 52.89 20,423/386.14 4,765/90.09 1,581/29.89 131/2.48 47/0.89 31/0.59 26,978/510.08 10 17.99 7,725/429.41 1,827/101.56 742/41.25 45/2.50 12/0.67 11/0.61 10,362/575.99 Total 320.75 131,636/410.40 30,150/94.00 10,885/33.94 805/2.51 225/0.70 191/0.59 173,892/542.14 The number that before and after “/” represent SSRs abundance and relative abundance Table 1.
Abundance and relative abundance of SSRs in the genome of Chinese cabbage.
-
Chromosome
no.Mono-
nucleotideDi-nucleotide Tri-nucleotide 1 A/T(12977) AC/GT(168) AG/CT(952) AAC/GTT(150) AAG/CTT(313) AAT/ATT(206) ACC/GGT(86) ACG/CGT(22) C/G(314) AT/TA(1971) CG/GC(1) ACT/AGT(12) AGC/CTG(36) AGG/CCT(129) ATC/ATG(136) CCG/CGG(23) 2 A/T(12822) AC/GT(155) AG/CT(753) AAC/GTT(98) AAG/CTT(331) AAT/ATT(201) ACC/GGT(56) ACG/CGT(29) C/G(289) AT/TA(2064) CG/GC(2) ACT/AGT(18) AGC/CTG(18) AGG/CCT(117) ATC/ATG(147) CCG/CGG(20) 3 A/T(16090) AC/GT(208) AG/CT(1080) AAC/GTT(182) AAG/CTT(469) AAT/ATT(223) ACC/GGT(100) ACG/CGT(24) C/G(289) AT/TA(2136) CG/GC(0) ACT/AGT(34) AGC/CTG(46) AGG/CCT(162) ATC/ATG(217) CCG/CGG(27) 4 A/T(9505) AC/GT(127) AG/CT(653) AAC/GTT(89) AAG/CTT(242) AAT/ATT(140) ACC/GGT(68) ACG/CGT(11) C/G(238) AT/TA(1550) CG/GC(0) ACT/AGT(7) AGC/CTG(30) AGG/CCT(89) ATC/ATG(80) CCG/CGG(15) 5 A/T(12623) AC/GT(183) AG/CT(1001) AAC/GTT(125) AAG/CTT(314) AAT/ATT(221) ACC/GGT(74) ACG/CGT(11) C/G(371) AT/TA(1898) CG/GC(1) ACT/AGT(24) AGC/CTG(37) AGG/CCT(138) ATC/ATG(140) CCG/CGG(22) 6 A/T(13546) AC/GT(167) AG/CT(1033) AAC/GTT(138) AAG/CTT(348) AAT/ATT(235) ACC/GGT(63) ACG/CGT(21) C/G(403) AT/TA(1992) CG/GC(0) ACT/AGT(30) AGC/CTG(34) AGG/CCT(113) ATC/ATG(151) CCG/CGG(21) 7 A/T(12388) AC/GT(146) AG/CT(882) AAC/GTT(126) AAG/CTT(345) AAT/ATT(175) ACC/GGT(57) ACG/CGT(5) C/G(245) AT/TA(1885) CG/GC(1) ACT/AGT(18) AGC/CTG(36) AGG/CCT(87) ATC/ATG(142) CCG/CGG(12) 8 A/T(11168) AC/GT(119) AG/CT(799) AAC/GTT(90) AAG/CTT(295) AAT/ATT(162) ACC/GGT(74) ACG/CGT(11) C/G(220) AT/TA(1630) CG/GC(1) ACT/AGT(16) AGC/CTG(33) AGG/CCT(99) ATC/ATG(107) CCG/CGG(9) 9 A/T(19818) AC/GT(262) AG/CT(1383) AAC/GTT(161) AAG/CTT(505) AAT/ATT(269) ACC/GGT(116) ACG/CGT(26) C/G(605) AT/TA(3119) CG/GC(1) ACT/AGT(20) AGC/CTG(59) AGG/CCT(164) ATC/ATG(238) CCG/CGG(23) 10 A/T(7552) AC/GT(106) AG/CT(655) AAC/GTT(95) AAG/CTT(245) AAT/ATT(129) ACC/GGT(45) ACG/CGT(14) C/G(173) AT/TA(1066) CG/GC(0) ACT/AGT(19) AGC/CTG(23) AGG/CCT(74) ATC/ATG(87) CCG/CGG(11) Table 2.
Amounts of different SSR repeat motifs in the genome of Chinese cabbage.
Figures
(3)
Tables
(2)