-
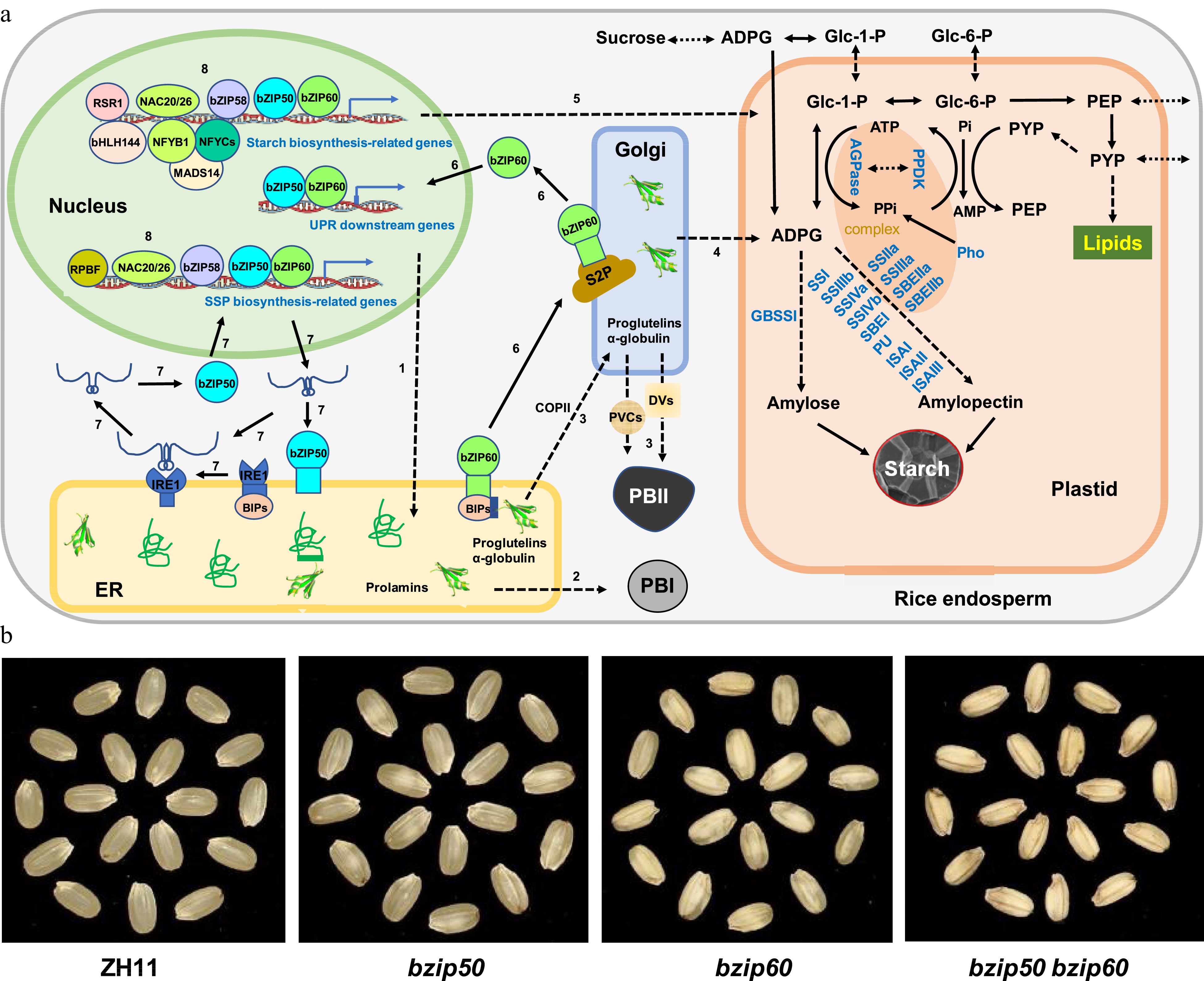
Figure 1.
Coordination of SSP and starch biosynthesis by the UPR pathways in rice endosperm. (a) Regulation of SSP and starch accumulation. Nascent polypeptides of SSPs with N-terminal signal peptide sequences are folded in the ER lumen (1), the properly folded prolamins are secreted and matured in PBI (2), while properly folded proglutelins and ɑ-globulin are exported from ER into Golgi via COPII vesicles and then to PBII mediated by DVs and PVCs (3). In contrast, proteins related to starch biosynthesis are transported either from Golgi to plastid (4) or from cytosol to plastid (5) to regulate starch biosynthesis. Two major UPR pathways, bZIP60-S2P and bZIP50-IRE1, are activated when ER protein homeostasis is disrupted in rice plants. Under such conditions, the ER membrane-associated transcription factor bZIP60 is disassociated with the chaperones BiPs, which results in the ER-to-Golgi transporting and processing of bZIP60 by Golgi-localized proteases such as S2P (6), the activated bZIP60 enters the nucleus and induces the expression of UPR downstream genes as well as SSP and starch biosynthesis-related genes (6). On the other hand, instead of encoding an ER membrane-associated transcription factor bZIP50, the bZIP50 mRNA is unconventionally spliced by the activated ER protein IRE1, leading to the open reading frame shift and a newly translated protein without any obvious transmembrane domain (7). The activated bZIP50 enters the nucleus to play a similar role as bZIP60 (7). Some other transcription factors, such as bZIP58 and NAC20/26, regulate the expression of genes involved in both SSP and starch biosynthesis (8). For protein abbreviations, see the text. (b) Phenotypic analysis. Wild-type (ZH11), single mutant of bZIP50 (bzip50) or bZIP60 (bzip60), and their double mutant (bzip50 bzip60) are grown under normal growth conditions and their dehulled seeds are photographed at maturity.
Figures
(1)
Tables
(0)