-
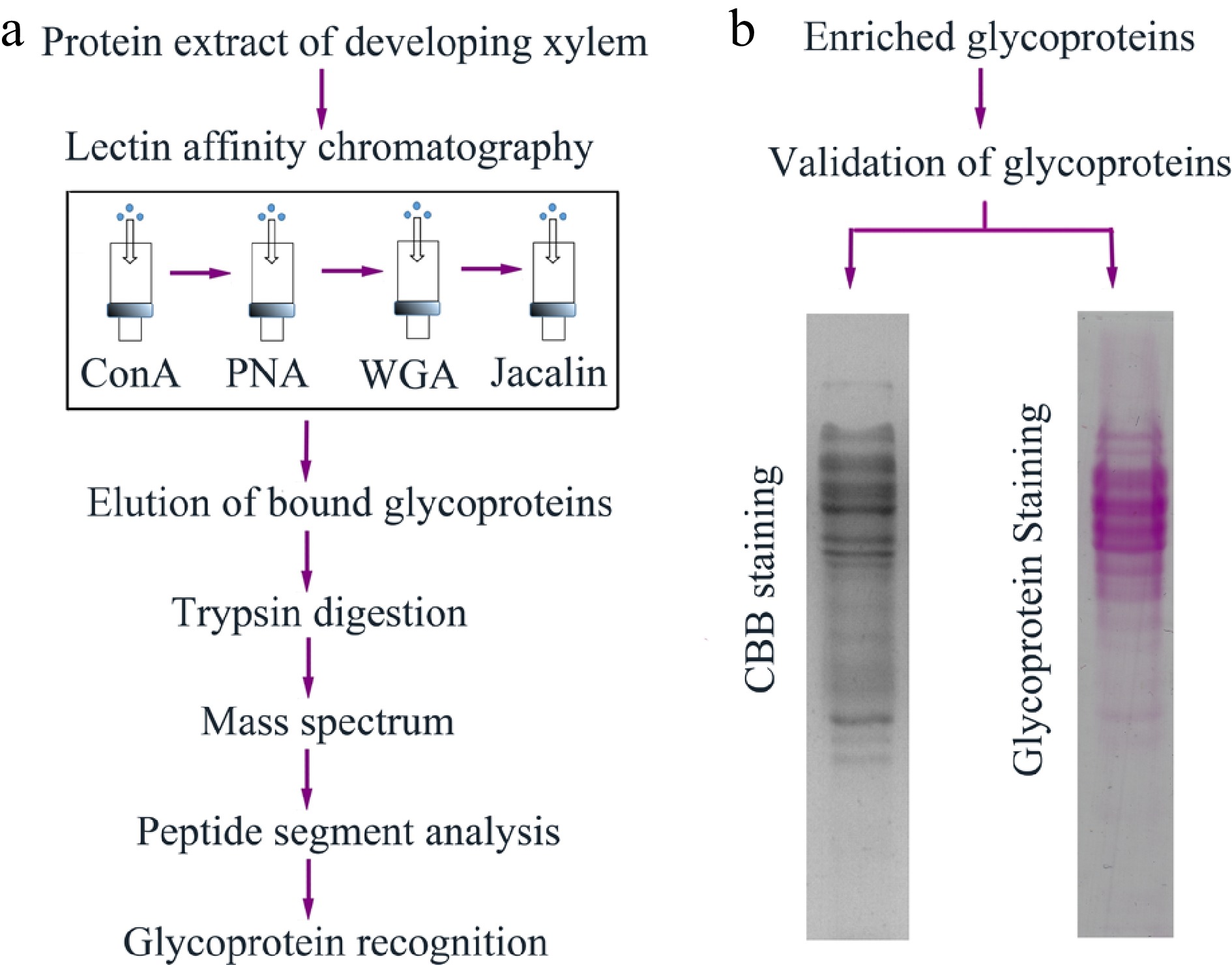
Figure 1.
The workflow of glycoprotein enrichment and identification in poplar developing xylem. (a) Enrichment and recognition of the glycoproteins. Crude proteins were extracted from poplar developing xylem, and the glycoproteins were bound to ConA, PNA, WGA and jacalin lectin affinity columns. The eluted glycoproteins were digested by trypsin and the peptide segments were further recognized through mass spectrum analysis. (b) Validation of the eluted glycoproteins. The glycoproteins separated on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels were stained using Coomassie brilliant blue (CBB) staining and Pierce Glycoprotein Staining kit (detecting sugar moieties of the glycoproteins), respectively.
-

Figure 2.
Verification of N- and/or O-glycosylation in PtSOD and HAD proteins. (a), (b) Analysis of PtSOD or PtHAD gene expression by RT-PCR in their transgenic Arabidopsis plants. AtActin2 serves as a control gene. (c), (d) Western blot analysis of PtSOD1-Flag and HAD-Flag protein levels in corresponding transgenic plants usig anti-FLAG antibody. Coomassie brilliant-stained Rubsico large subunit proteins indicate the loading amount of each sample on SDS-PAGE gels as control. (e), (f) Migration analysis of PtSOD and PtHAD proteins on SDS-PAGE gels. Protein extracts with/without the digestion of PNGase F (N) or O-glycosidase (O) were separated on 10% SDS-PAGE gels and followed by immunoblotting with anti-FLAG antibody.
-
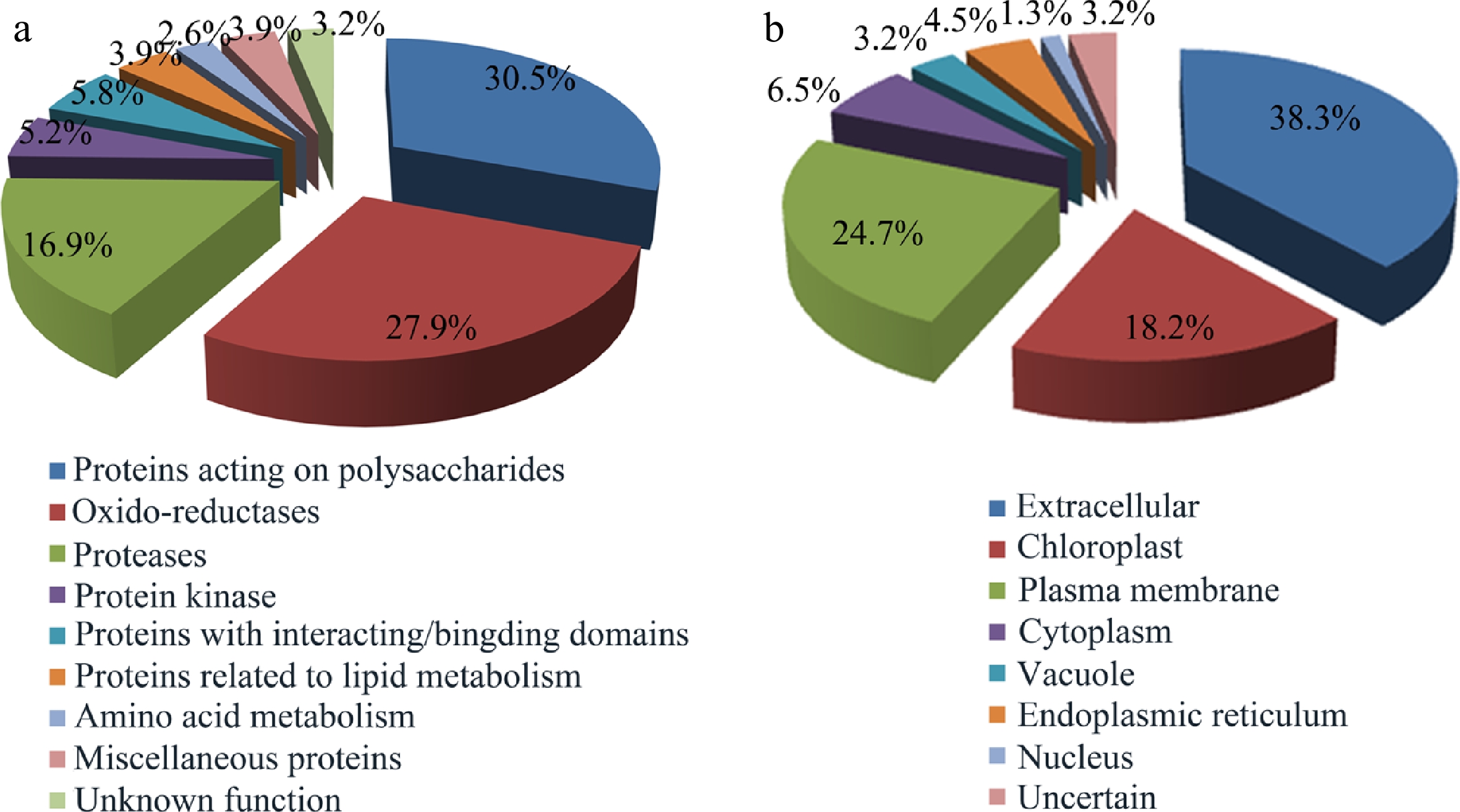
Figure 3.
Functional classification and localization of the identified glycoproteins. (a) Functional classification of glycoproteins identified from Populus developing xylem. Please refer to Supplemental Table S1 for detailed analysis. (b) Subcellular localization of the identified glycoproteins predicted by Plant-mPLoc, ngLOC, ProtComp9.0, WoLF PSORT and YLOC.
-
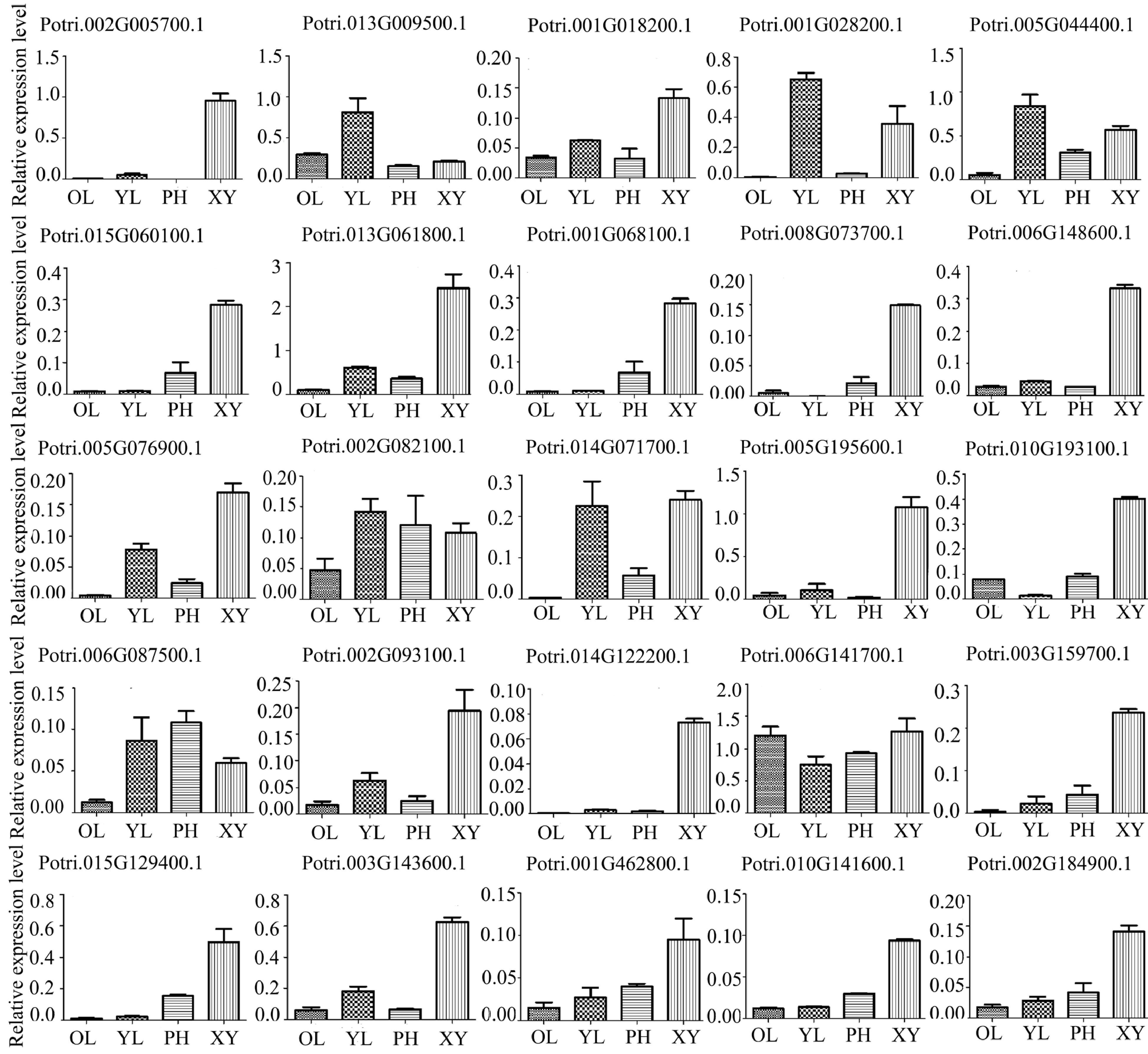
Figure 4.
Expression profiles of 25 genes encoding the glycoproteins in different poplar tissues using RT-qPCR analysis. Different tissues included phloem (PH), xylem (XY), young leaf (YL) and mature leaf (OL). The expression of PtActin2 was used as an internal control. Data are means ± standard error of three technical replicate results.
-
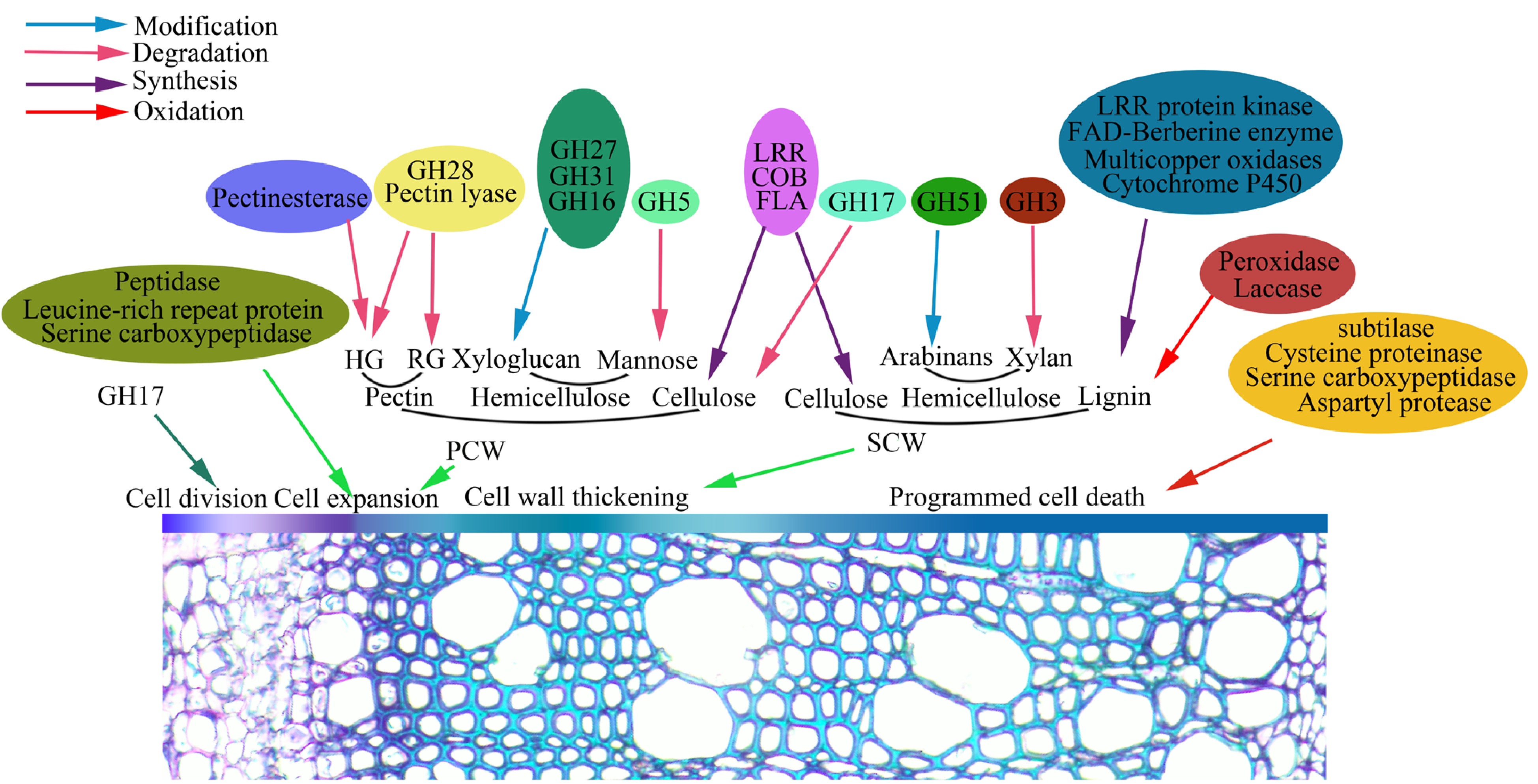
Figure 5.
Identified glycoproteins are proposed to be involved in wood formation that includes cambia cell division, cell differentiation and expansion, SCW synthesis and PCD.
-
Protein name Gi number Phytozome
accession no.Peptides Score Cov % N-/O-linked sites Proteins acting on polysaccharides (47) GH3 Beta-xylosidase 222845455 Potri.001G206800 9 432 16 Y/Y GH3 Beta-xylosidase 222846715 Potri.001G089100 6 188 10 Y/Y GH3 Beta-xylosidase 222861083 Potri.014G122200 6 285 11 Y/N GH3 Beta-xylosidase 222844484 Potri.002G197200 1 76 1 Y/N GH3 Beta-glucosidase 222852772 Potri.007G114300 1 48 1 Y/Y GH5 Mannan endo-1,4-beta-mannosidase 222855167 Potri.006G109900 2 86 6 Y/Y GH16 Xyloglucan endo-transglycosylase 118481141 Potri.003G159700 6 483 26 Y/N GH16 Xyloglucan endo-transglycosylase 124109187 Potri.001G071000 6 414 28 Y/N GH16 Xyloglucan endo-transglycosylase 222857312 Potri.013G005700 2 203 9 Y/Y GH17 Glucan endo-1,3-beta-glucosidase 222850378 Potri.009G076500 3 86 8 Y/Y GH17 Glucan endo-1,3-beta-glucosidase 222858075 Potri.013G059700 12 1333 39 Y/N GH17 Glucan endo-1,3-beta-glucosidase 222862285 Potri.019G032900 10 1131 29 Y/Y GH17 Glucan endo-1,3-beta-glucosidase 222873604 Potri.018G150400 7 671 20 Y/Y GH 27 Alpha-galactosidase 222862356 Potri.019G056700 1 64 2 Y/Y GH28 polygalacturonase-like 222843096 Potri.002G162400 5 446 20 Y/N GH28 polygalacturonase 222863392 Potri.010G005500 3 167 9 Y/N GH28 polygalacturonase 222838571 Potri.008G211500 3 160 10 Y/N GH28 polygalacturonase 222861707 Potri.014G112100 2 134 7 Y/N GH28 polygalacturonase 222837934 Potri.003G131700 1 125 3 Y/Y GH28 polygalacturonase 222860156 Potri.011G159000 2 84 4 Y/N GH28 polygalacturonase 222843280 Potri.002G186900 2 60 6 Y/N GH28 polygalacturonase 222867323 Potri.016G051200 2 53 4 Y/Y GH31 Glucan 1,3-alpha-glucosidase 222853440 Potri.007G100000 10 287 13 Y/N GH31 Glucan 1,3-alpha-glucosidase 222856503 Potri.005G069000 4 97 6 Y/Y GH32 beta-fructofuranosidase 222868827 Potri.015G127100 2 46 5 Y/Y GH38 alpha-mannosidase 222843486 Potri.002G238200 14 859 19 Y/N GH38 alpha-mannosidase 222861848 Potri.014G143600 10 521 15 Y/N GH38 alpha-mannosidase 222859443 Potri.012G106500 5 374 7 Y/Y GH38 alpha-mannosidase 222859442 Potri.012G106400 1 50 7 N/Y GH51 Alpha-L-arabinofuranosidase 222853916 Potri.006G029900 2 269 5 Y/Y GH127 Beta-L-arabinofuranosidase 222845043 Potri.001G018200 5 154 7 Y/Y Alpha-fucosidase 222863630 Potri.010G047900 3 151 11 Y/N Xylose isomerase 222865922 Potri.T093900 13 998 37 N/N Pectinesterase 222861105 Potri.014G127000 2 285 6 Y/Y Pectinesterase 222844452 Potri.002G202600 1 263 3 Y/Y Pectin lyase 118488323 Potri.003G175900 2 71 6 Y/Y Acetylglucosaminyl transferase 222845138 Potri.001G068100 2 48 7 Y/N Glycopeptide N-glycosidase 222859921 Potri.011G109700 7 268 11 Y/Y Glucosidase II beta subunit 222872983 Potri.006G061600 1 74 4 Y/N Fasciclin-like arabinogalactan protein 222861509 Potri.014G071700 2 99 7 Y/Y Fasciclin-like arabinogalactan protein 47717933 Potri.015G129400 2 96 10 Y/Y Fasciclin-like arabinogalactan protein 118482997 Potri.012G127900 2 84 10 Y/Y Non-classical arabinogalactan protein 118482413 Potri.002G093100 1 82 10 N/Y Non-classical arabinogalactan protein 118481929 Potri.004G044700 1 79 6 Y/Y COBRA-like protein 118485798 Potri.010G001100 5 232 12 Y/Y COBRA-like protein 118488472 Potri.015G060100 4 145 11 Y/N COBRA-like protein 118482010 Potri.015G060000 1 50 2 Y/Y Oxido-reductases (43) Multicopper oxidase, SKU5-like protein 222871142 Potri.001G120300 13 1097 36 Y/Y Multicopper oxidase, SKU5-like protein 222840952 Potri.003G112700 12 944 34 Y/Y Multicopper oxidase, SKS1-like protein 222859558 Potri.012G126400 5 232 14 Y/Y Multicopper oxidase, SKS1-like protein 222868828 Potri.015G127200 3 167 8 Y/Y Multicopper oxidase, SKS4-like protein 118487967 Potri.004G010100 1 182 2 Y/N Multicopper oxidase 222857214 Potri.005G247700 10 1031 30 Y/N Multicopper oxidase 222842395 Potri.002G013700 14 997 27 Y/N Multicopper oxidase 222853065 Potri.007G038300 10 844 29 Y/Y Multicopper oxidase 118488761 Potri.001G219300 9 526 26 Y/Y Multicopper oxidase 222849177 Potri.004G180500 12 470 26 Y/N Multicopper oxidase 222843342 Potri.002G227600 2 194 6 Y/Y Multicopper oxidase 222855045 Potri.006G087500 4 142 9 Y/Y Multicopper oxidase 222844867 Potri.001G000600 1 109 2 Y/N Multicopper oxidase 222849246 Potri.009G159700 4 56 9 Y/Y Laccase 222852007 Potri.010G183500 6 1201 16 Y/Y Laccase 222852006 Potri.008G073700 9 1113 23 Y/Y Laccase 222864170 Potri.010G183500 5 1006 13 Y/Y Laccase 222864171 Potri.010G183600 5 639 13 Y/N Laccase 222854184 Potri.006G096900 3 95 7 Y/Y Laccase 222849832 Potri.009G034500 3 79 7 Y/N Laccase 222850532 Potri.009G042500 1 65 2 Y/Y Laccase 222846554 Potri.001G054600 1 64 3 Y/N Laccase 3805960 Potri.010G193100 1 46 2 Y/Y peroxidase 115345276 Potri.003G214700 5 514 21 Y/N Peroxidase 118487605 Potri.005G195600 3 55 12 Y/Y FAD-Berberine enzyme 545aa 222860154 Potri.011G158700 7 323 15 Y/Y FAD-Berberine enzyme 222846288 Potri.001G462800 5 298 10 Y/N FAD-Berberine enzyme 222833370 Potri.006G128900 3 223 5 Y/N FAD-Berberine enzyme 222872123 Potri.011G159600 2 175 3 Y/N FAD-Berberine enzyme 222846286 Potri.001G462600 2 175 2 Y/Y FAD-Berberine enzyme 222858409 Potri.012G034700 2 175 3 Y/N FAD-Berberine enzyme 222834675 Potri.011G160300 2 173 5 Y/N FAD-Berberine enzyme 222872175 Potri.011G161500 2 89 4 Y/N FAD-Berberine enzyme 222846302 Potri.001G464700 1 83 2 Y/Y FAD-Berberine enzyme 222872118 Potri.011G161400 1 80 2 Y/N FAD-Berberine enzyme 222847838 Potri.001G470100 1 80 2 N/Y FAD-Berberine enzyme 222860155 Potri.011G158800 1 80 2 Y/Y Protein disulfide isomerase 222842706 Potri.002G082100 23 3116 50 Y/Y Protein disulfide isomerase 118485031 Potri.009G013600 14 648 29 Y/N Protein disulfide isomerase 222846968 Potri.001G183500 2 55 5 Y/Y Chitinase-like 118481023 Potri.010G141600 2 65 9 Y/Y Cytochrome P450 222868639 Potri.015G085800 1 56 N/Y Cu/Zn superoxide dismutase 4102861 Potri.005G044400 1 63 10 Y/Y Proteases (26) Aspartyl protease 118482048 Potri.001G028200 5 244 15 Y/Y Aspartyl protease 439aa 222847473 Potri.001G306200 1 90 3 Y/N Serine carboxypeptidase 222849960 Potri.009G003100 2 129 7 Y/N Serine carboxypeptidase 222850469 Potri.009G055900 4 88 8 Y/Y Serine carboxypeptidase S28 222854432 Potri.006G207900 3 499 10 Y/Y Serine carboxypeptidase S28 222836225 Potri.007G015400 6 428 14 Y/N Serine carboxypeptidase S28 118487876 Potri.007G015300 5 418 13 Y/N Serine carboxypeptidase S28 222853228 Potri.007G008100 4 224 12 Y/N Subtilase family protein 222860749 Potri.011G155400 4 269 7 Y/N Subtilase family protein 222875305 Potri.001G440300 5 243 9 Y/N Subtilase family protein 222848475 Potri.004G173900 1 81 1 Y/Y Subtilase family protein 222854095 Potri.006G076200 3 76 5 Y/Y Peptidase M20/M25/M40 222863686 Potri.010G076100 11 1315 33 Y/N Peptidase M20/M25/M40 118486005 Potri.009G169300 12 912 41 N/Y Peptidase M20/M25/M40 222837797 Potri.004G208100 4 526 12 N/Y Peptidase M20/M25/M40 222842722 Potri.002G085400 5 205 16 Y/Y Peptidase M20/M25/M40 222840651 Potri.003G045200 2 131 5 Y/N Peptidase M28 family 222855209 Potri.006G153300 4 178 10 Y/N Cysteine proteinase 222856445 Potri.005G256000 4 320 16 Y/N Cysteine proteinase 222843627 Potri.002G005700 4 223 21 Y/N Cysteine proteinase 222837653 Potri.004G207600 3 212 12 Y/N Cysteine proteinase 118482340 Potri.006G141700 1 59 3 Y/N Proteinase inhibitor 118485178 Potri.013G112800 3 56 39 N/Y Proteinase inhibitor 118482991 Potri.019G083300 2 51 20 N/Y Amidohydrolase family 222849678 Potri.009G067700 5 122 12 Y/Y Amidohydrolase family 222847228 Potri.001G273400 4 59 11 Y/Y Protein kinase (8) LRR protein kinase 222868332 Potri.016G144100 4 480 7 Y/Y LRR protein kinase 222853199 Potri.007G014700 3 231 4 Y/Y LRR protein kinase 222863806 Potri.010G103000 4 133 7 Y/Y LRR protein kinase 222854082 Potri.006G073900 4 110 6 Y/Y LRR protein kinase 222866571 Potri.018G107400 2 98 2 Y/Y LRR protein kinase 222852307 Potri.008G140500 1 45 1 Y/Y LRR protein kinase 222856570 Potri.005G083000 3 45 3 Y/Y LRR protein kinase 222852450 Potri.008G176900 1 47 2 Y/N Proteins with interacting/binding domains (9) Leucine-rich repeat protein 190897432 Potri.009G064300 9 663 42 Y/N Leucine-rich repeat protein 222853264 Potri.007G001000 5 348 12 Y/N Leucine-rich repeat protein 222854117 Potri.018G151000 2 132 4 Y/Y Leucine-rich repeat protein 222846498 Potri.001G017500 1 46 2 Y/N HSP70 family protein 222867185 Potri.016G019800 13 998 24 Y/N HSP70 family protein 222854802 Potri.006G022100 14 939 20 Y/Y HSP70 family protein 222841104 Potri.003G143600 1 62 2 N/Y Calreticulin family protein 222871704 Potri.005G015100 11 834 34 Y/Y Calreticulin family protein 118485765 Potri.013G009500 10 653 38 Y/Y Proteins related to lipid metabolism(6) Lipase/lipooxygenase 118483838 Potri.005G076900 5 287 40 Y/N Purple acid phosphatase 222865126 Potri.010G158400 3 119 7 Y/N Purple acid phosphatase 222851161 Potri.008G096000 3 109 6 Y/N HAD superfamily protein 222839124 Potri.004G232900 2 63 9 Y/N Type I phosphodiesterase 222855200 Potri.006G150900 2 160 7 Y/Y Type I phosphodiesterase 222872448 Potri.018G066600 1 79 3 Y/Y Amino acid metabolism (4) Amidase family protein 222869309 Potri.015G109400 4 180 12 Y/Y Methionine synthase 222850043 Potri.009G152800 2 56 4 Y/Y Methionine synthase 118483919 Potri.013G061800 2 56 4 Y/Y Cysteine desulfurase 222850426 Potri.009G066000 1 55 Y/Y Miscellaneous proteins (6) Germin-like protein 10 118482567 Potri.002G184900 4 623 19 Y/Y Cyclase family protein 222850275 Potri.009G097300 2 192 13 Y/N Cyclase family protein 118488222 Potri.001G301600 4 95 18 Y/Y Kelch repeat protein 222845394 Potri.001G178500 1 73 Y/N Nucleosome assembly protein 222854259 Potri.006G148600 1 56 3 Y/N Cupin domain protein 222858047 Potri.013G051600 1 55 7 Y/N Unknown function (5) Unknown protein (Duf642) 118486479 Potri.011G087500 4 124 16 Y/Y Unknown protein (Duf568) 118487890 Potri.002G249200 1 54 5 Y/Y Unknown protein (Duf2828) 222850304 Potri.009G091400 1 57 Y/Y Unknown protein 222846617 Potri.001G068800 1 80 6 Y/Y Unknown protein 118486279 Potri.019G076900 2 68 5 Y/Y Table 1.
Identification of the proteins enriched by lectin affinity from poplar developing xylem.
Figures
(5)
Tables
(1)