-
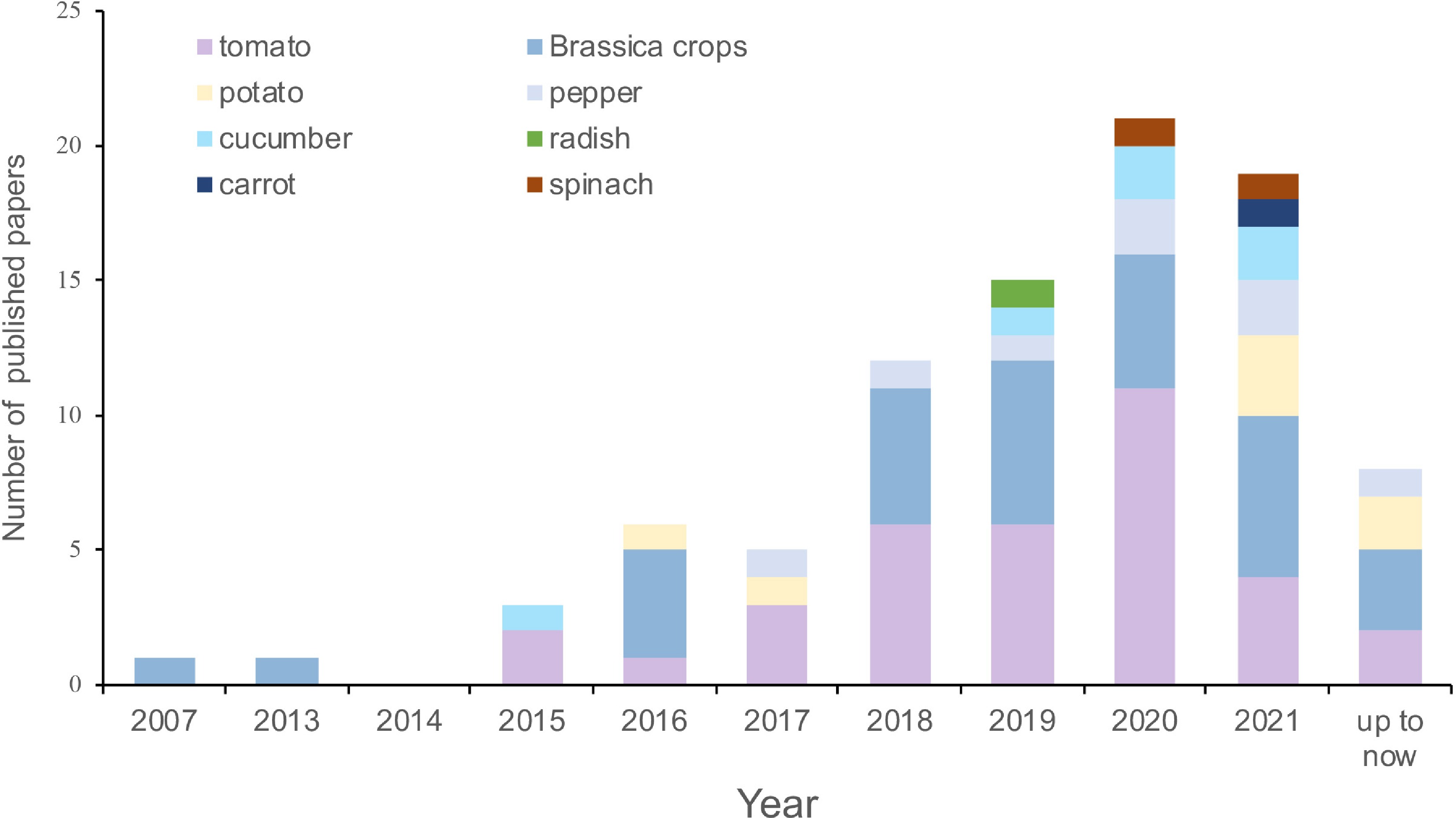
Figure 1.
Statistics on the published number of lnRNA-related papers of major vegetables.
-
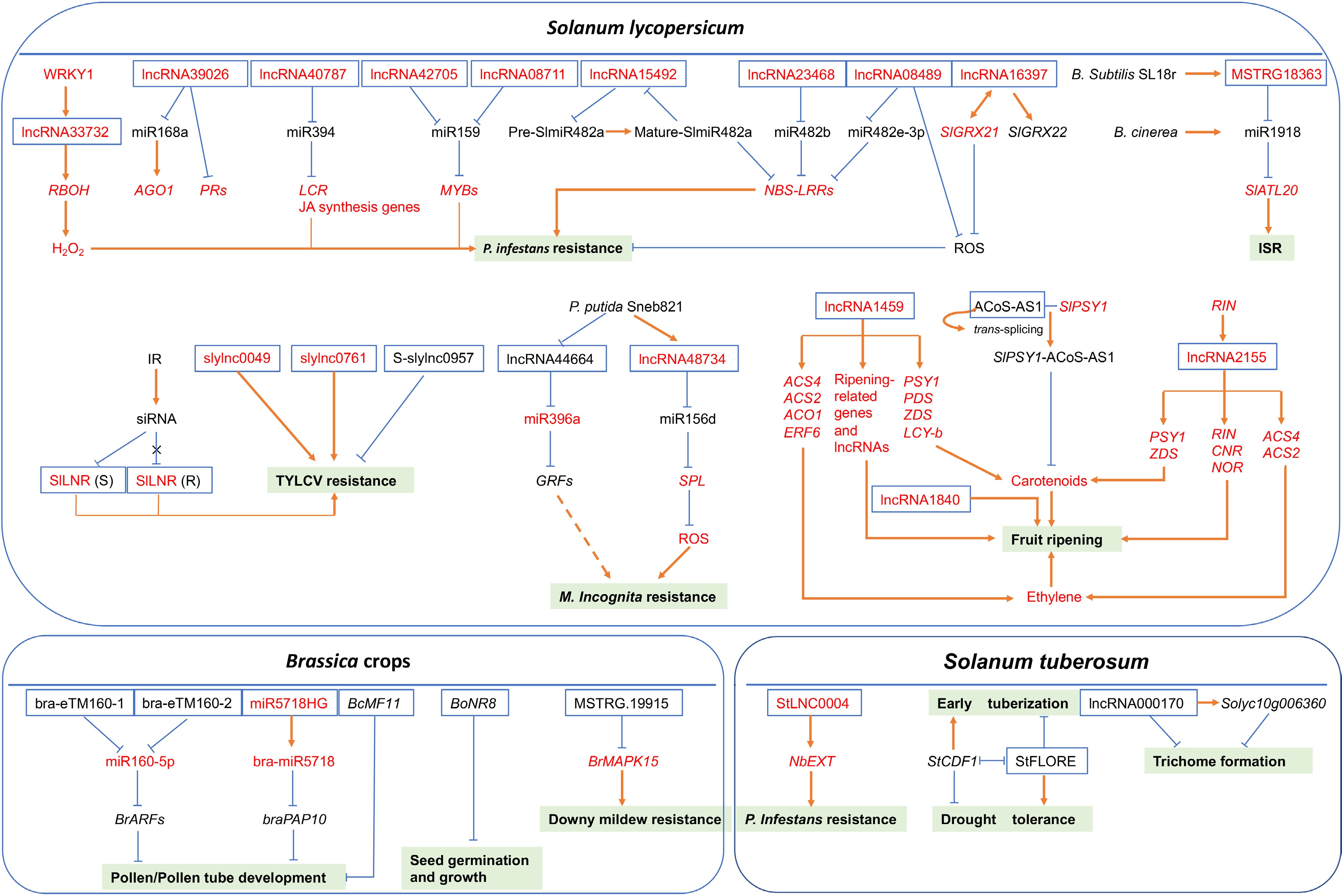
Figure 2.
The predictive model of regulatory mechanisms of lncRNAs with known functions under various developmental events or stress conditions in different vegetable crops. Full-line arrows represent positive regulatory interactions, blunt-ended bars represent negative regulation and dotted-line arrow indicates that the regulatory mechanism is unclear. White boxes with blue lines represent lncRNAs, light green boxes represent different developmental events or stresses. The letters in red denote positive regulators, while the letters in black denote negative regulators.
-
Roles Species Pathways Approaches LncRNAs
numberDE-LncRNAs number Ref. Growth and development Solanum lycopersicum Fruit ripening RNA-seq − 378 [41] Solanum lycopersicum Fruit ripening ssRNA-seq 3,679 677 [61] Solanum lycopersicum Fruit expansion and
ripeningssRNA-seq 17,674 tissue- and stage-dependent [60] Solanum lycopersicum RIN target lncRNAs; fruit ripening ChIP-seq & RNA-seq 187 − [38] Solanum lycopersicum Fruit ripening integrate 134 data sets 79,322 tissue- and stage- specificity [22] Capsicum chinense Jacq. Fruit ripening RNA-seq 20,563 1,1826 [64] Capsicum annuum Fruit ripening RNA-seq 11,999 366 [65] Capsicum annuum Fruit development ssRNA-seq 2,505 1,066 [66] Brassica rapa Vernalization RNA-seq 1,961 254 [42] Brassica rapa var. pekinensis Vernalization RNA-seq 2,088 549 [68] Brassica
campestris ssp. pekinensisVernalization ssRNA-seq 1,858 151 [69] Brassica rapa Pollen development RNA-seq 12,051 14 [43] Brassica rapa ssp. pekinensis Anther development SMRT 407 − [34] Brassica campestris ssp. pekinensis Anther development RNA-seq 2,384 1,344 [72] Brassica rapa ssp. pekinensis Cytoplasmic male sterility RNA-seq 3,312 529 [74] Brassica
campestrisMale sterile RNA-seq 13,879 361 [73] Capsicum annuum Cytoplasmic male sterilitye RNA-seq 10,655 1,137 [75] Solanum lycopersicum Sperm cell lineage
developmentssRNA-seq 31,931 cell/tissue-type specificity [76] Capsicum annuum Anthocyanin biosynthesis ssRNA-seq − 172 [44] Solanum tuberosum Anthocyanin Biosynthesis ssRNA-seq 4,376 1,421 [80] Solanum tuberosum Anthocyanin Biosynthesis RNA-seq 1,072 6 [81] Daucus carota Anthocyanin biosynthesis RNA-seq 7,288 639 [82] Solanum lycopersicum Trichome formation ssRNA-seq 1,303 196 [83] Solanum tuberosum Potato tuber sprouting RNA-seq 3,175 723 [87] Spinacia oleracea Flowering RNA-seq 1,141 111 [89] Spinacia oleracea Sex differentiation PacBio Iso-seq & RNA-seq 500 42 [37] Growth and development Brassica napus Oil
biosynthesisssRNA-seq & RNA-seq datasets 8,905 13 [90] Brassica oleracea
var. capitataCuticular wax
biosynthesisRNA-seq 4,459 148 [91] Abiotic stress Solanum lycopersicum Drought response RNA-seq 521 244 [45] Solanum lycopersicum drought-response RNA-seq 67,770 3,053 [103] Brassica napus Drought response RNA-seq - 477/706 [104] Solanum tuberosum Drought response NGS & SMRT 3,445 − [105] Brassica rapa Heat response ssRNA-seq 4,594 1,686 [46] Brassica juncea Heat and drought response RNA-seq 7,613 1,614 [107] Brassica rapa Heat response RNA-seq 18,253 1,229 [15] Brassica rapa ssp. pekinensis Heat response RNA-seq 278 65 [106] Brassica rapa ssp. chinensis
(NHCC)Cold and heat response RNA-seq 10,001 2,236 [108] Cucumis sativus Heat response RNA-seq 2,085 108 [109] Raphanus sativus Heat response ssRNA-seq − 169 [110] Solanum lycopersicum Chilling injury RNA-seq 1,411 239 [47] Capsicum annuum Chilling injury RNA-seq 9,848 380 [111] Solanum lycopersicum Fruit cracking RNA-seq 2,508 − [112] Solanum pennellii and M82 Salt response ssRNA-seq 1,044 154/137 [48] Cucumis sativus Waterlogging response RNA-seq 3,738 922/514/1,476/
1,270[115] Cucumis sativus Phosphate-deficiency
responsessRNA-seq 14,277 22 [121] Brassica
napusCadmium toxic response ssRNA-seq 5,038 301 [126] Biotic stress
Solanum tuberosum Phytophthora infestans resistance RNA-seq 2,857 133 [49] Solanum lycopersicum Phytophthora
infestans resistanceRNA-seq 28,256 688 [130] Solanum lycopersicum L3708 Phytophthora
infestans resistanceRNA-seq 9,011 196 [131] Solanum lycopersicum TYLCV resistance ssRNA-seq 1,565 529 [50] Solanum lycopersicum TYLCV resistance ssRNA-seq 2,056 345 [138] Biotic stress
Solanum lycopersicum Bacillus subtilis SL18r-induced tomato resistance against Botrytis cinerea RNA-seq − 55/34/15 [51] Solanum lycopersicum Pseudomonas putida Sneb821- induced tomato resistance against Meloidogyne incognita RNA-seq 3,371 78 [140] Solanum lycopersicum Pst resistance RNA-seq 2,609 Different in each comparison [52] Solanum lycopersicum PSTVd resistance RNA-seq 6,726 44 [141] Brassica campestris ssp.chinensis Makino Plasmodiophora brassicae resistance RNA-seq 1,492 114 [143] Brassica napus Plasmodiophora brassicae resistance ssRNA-seq 4,558 530 [144] Brassica rapa ssp. pekinensis Downy mildew resistance RNA-seq 3,711 − [145] Brassica napus Sclerotinia sclerotiorum resistance RNA-seq 3,181 931 [142] Brassica rapa Fusarium oxysporum
resistanceqPCR [146] Capsicum
annuumPhytophthora
capsica resistanceRNA-seq 2,388 607 [147] Solanum tuberosum Pectobacterium carotovorum resistance ssRNA-seq 1,113 559 [148] Solanum tuberosum Potato Virus Y resistance and heat stress RNA-seq 4,007 421 [149] Cucumis sativus Powdery mildew resistance ssRNA-seq 12,903 119 [150] Others Solanum lycopersicum Ethylene signaling RNA-seq 397 12 [151] Solanum
pimpinellifolium LA1589,
S. lycopersicum Heinz1706Lycopersicon specificity ssRNA-seq 413/709 92/161 [152] Brassica napus, B. oleracea and
B. rapaSpecies divergence RNA-seq 1,885/1,910/
1,299186 /157/161 [153] Capsicum chinense Heterosis effect ssRNA-seq 2,525 1,932/ 593 [154] Cucumis hytivus Allopolyploidization RNA-seq 2,206 1,328 [155] Brassica rapa, B. carinata, and
B. hexaploidPolyploidization RNA-seq 2,725/1,672/
2,810725 [156] Table 1.
List of long non-coding RNAs (lncRNAs) identified in major vegetable crops.
-
Species LncRNA name Biological functions Interaction targets References Solanum lycopersicum lncRNA000170 Trichome formation Solyc10g006360 [83] lncRNA1459, lncRNA1840 Fruit ripening − [61, 62] lncRNA2155 Fruit ripening RIN [38] ACoS-AS1 Trans-splicing; carotenoids biosynthesis SlPSY1 [63] lncRNA33732 Resistance to Phytophthora infestans RBOH [132] lncRNA16397 Resistance to Phytophthora infestans SlGRX21, SlGRX22 [130] lncRNA15492 Resistance to Phytophthora infestans Sl-miR482a [133] lncRNA08489 Resistance to Phytophthora infestans miR482e-3p [134] lncRNA23468 Resistance to Phytophthora infestans miR-482b [135] lncRNA39026 Resistance to Phytophthora infestans miR-168a [136] lncRNA40787 Resistance to Phytophthora infestans miR394 [137] lncRNA42705, lncRNA08711 Resistance to Phytophthora infestans miR159 [131] slylnc0049, slylnc0761 Resistance to TYLCV − [50] S-slylnc0957 Resistance to TYLCV − [138] SlLNR1 Resistance to TYLCV − [139] MSTRG18363 Bacillus subtilis SL18r-induced tomato resistance against Botrytis cinerea miR1918 [51] lncRNA44664, Pseudomonas putida Sneb821- induced tomato resistance against Meloidogyne incognita miR396 [140] lncRNA48734 Pseudomonas putida Sneb821- induced tomato resistance against Meloidogyne incognita miR156 [140] Brassica oleracea BoNR8 Seed germination; root and silique growth − [21] Brassica rapa bra-eTM160-1, bra-eTM160-2 Pollen development miR160-5p [43] Brassica rapa ssp. pekinensis MSTRG.19915 Resistance to downy mildew BrMAPK15 [145] Brassica campestris bra-miR5718HG Pollen tube growth miR5718 [73] BcMF11 Pollen development; male fertility − [39, 40] Solanum tuberosum StFLORE Tuber development; drought response StCDF1 [88] StLNC0004 Resistance to Phytophthora infestans NbEXT [49] Table 2.
Summary of functionally validated lncRNAs in major vegetables.
Figures
(2)
Tables
(2)