-
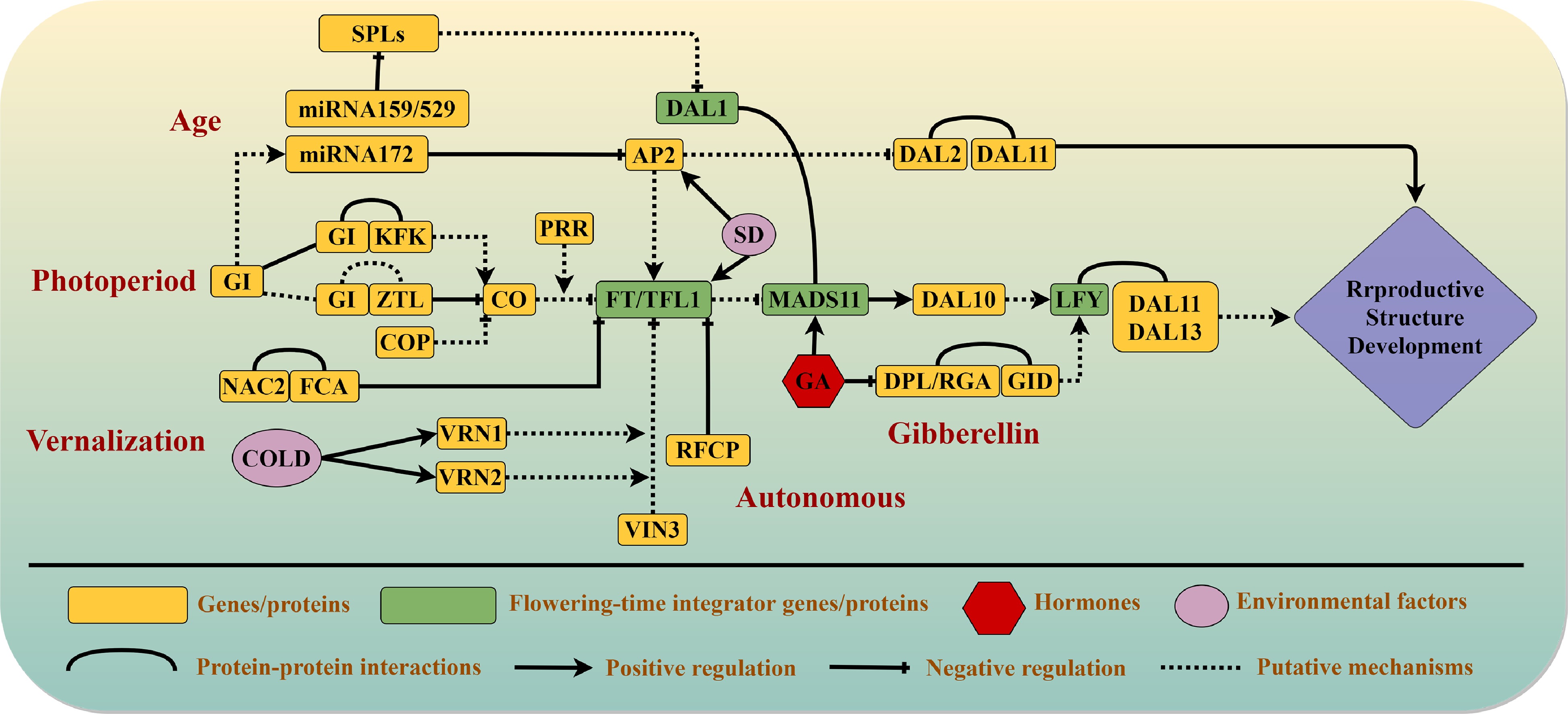
Figure 1.
Current understanding of flowering pathways in conifers. Five interdependent pathways control the reproductive transition in conifers: the vernalization, photoperiod, autonomous, gibberellin, and aging pathways. Arrows indicate promotion, blunt-ended lines indicate genetic inhibition, and curves indicate protein–protein interactions. Solid lines denote interactions that are supported by experimental evidence, whereas dashed lines denote proposed interactions. Genes that act as major regulators in different pathways are written in green blocks. Environmental factors are represented by pink ellipses, and hormones involved in reproductive growth are represented by red hexagons.
-
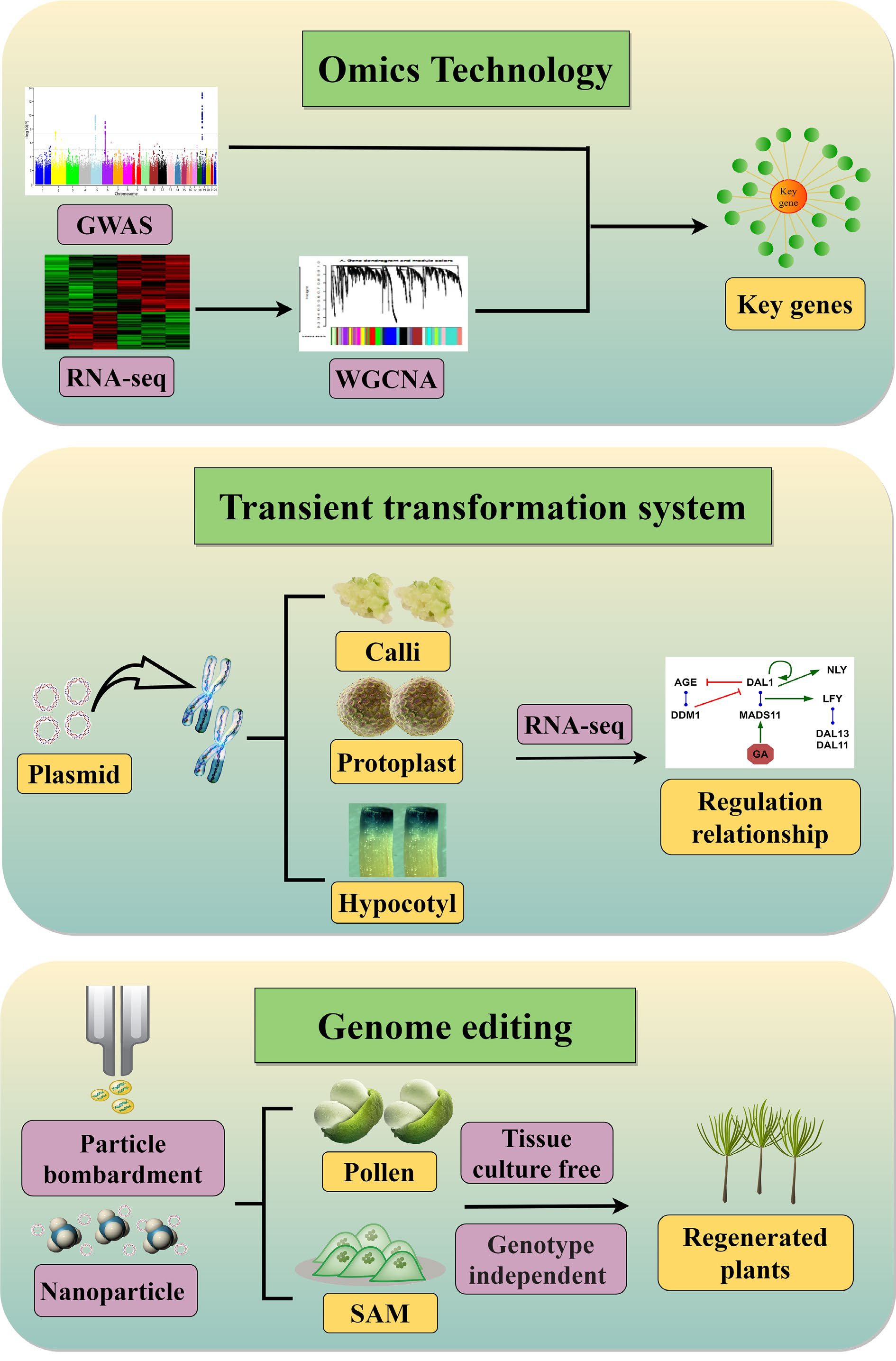
Figure 2.
Strategies for identification and characterization of conifer genes and their regulatory relationships. Omics technologies combine GWAS and WGCNA based on RNA-seq data to identify key genes that determine important traits. Transient transformation systems overcome restrictions on genetic transformation, enabling integration of target plasmids into conifer chromosomes to produce functional proteins. A genetic regulatory network can then be constructed from RNA-seq data. The aim of genome editing is to precisely modify target genes or regulatory elements in conifers. Tissue culture–free delivery systems include delivery via plant germline or meristematic cells and nanotechnology-based delivery systems.
Figures
(2)
Tables
(0)