-
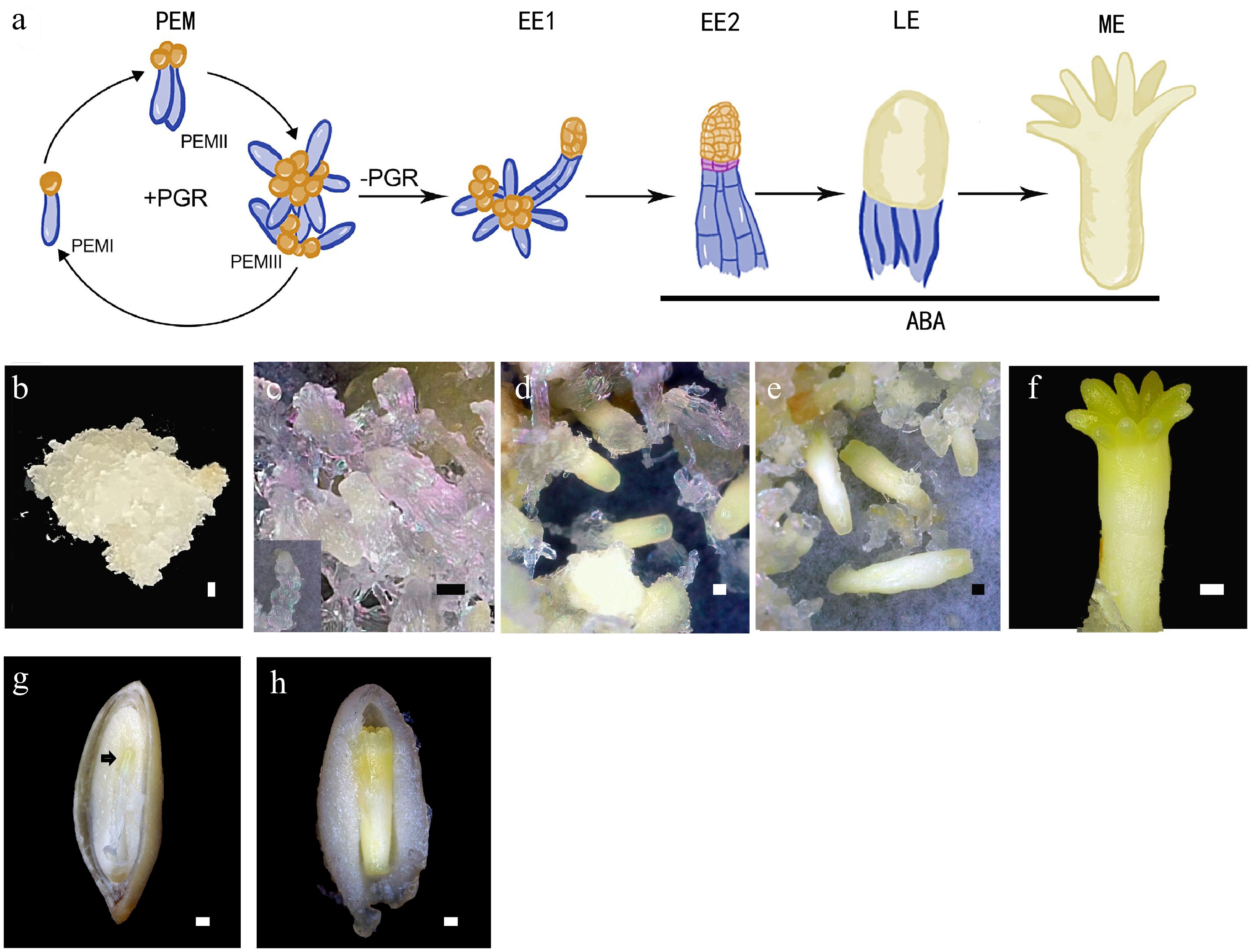
Figure 1.
Conifer embryo development represented by Picea abies. (a) Schematic representation of the developmental stages of somatic embryo development. (b)−(f) Somatic embryo development process: (b) proembryogenic mass (PEM); (c) cultures after one week on maturation medium, insert presents an early somatic embryo (EE); (d) culture contains late embryos (LEs); (e) culture contains maturing and (f) matured somatic embryo (ME). (g), (h) Dissected seeds to show (g) the early zygotic embryo, indicated by the black arrow, and (h) the maturing zygotic embryo. Bar = 500 μm.
-
Gene family Gene Description References LRR-RLKs SOMATIC EMBRYOGENESIS RECEPTOR-
LIKE KINASE 1-5 (SERKs)Transmembrane proteins; involved in signal transduction and have been strongly associated with somatic embryogenesis and apomixis in a number of plant species. [23] AP2/ERF BABYBOOM (BBM) Tanscription factor; activates LEC1-ABI3-FUS3-LEC2 network to induce somatic embryogenesis. [24] EMBRYOMAKER (EMK/AIL5 ) Tanscription factors; promote the formation of somatic embryo on cotyledons. [25] WOUND INDUCED DEDIFFERENTIATION1 (WIND1) Tanscription factor; controls cell dedifferentiation in Arabidopsis and functions as a key molecular switch for plant cell dedifferentiation. [26,27] B3-AFL LEAFY COTYLEDON 1 (LEC1) Tanscription factor; promote somatic embryo development in vegetative organs. [28] LEC1-LIKE (L1L) Tanscription factor; promote somatic embryo development in vegetative organs. [29] LEAFY COTYLEDON 2 (LEC2) Tanscription factor; activates the expression of embryonic traits in vegetative tissues. [28] ABSCISIC ACID INSENSITIVE 3 (ABI3)/VIVI PAROUS (VP1) Transcript factor; regulates embryo-specific ABA-induced genes. [30] FUSCA3 (FUS3) Transcription factor; promotes embryogenesis by regulating synthesis of storage proteins and lipids. [31] VP1/ABI3-LIKE (VAL) Transcription factor; repress plant embryo development. [32] WOX WUSCHEL (WUS) Transcription factor; a central player in stem cell maintenance in the SAM. [33] WUSCHEL-related homeobox (WOX) 2 Transcription factor; promotes apical embryonic cell division. [34] WOX 5 Transcription factor; a central player in stem cell maintenance in the SAM. [35] WOX 8 and WOX9 WOX8 and WOX9 functionally overlap in promoting basal embryonic cell division. [34] NAC CUP SHAPED COTYLEDONS 1-3 (CUCs) CUP SHAPED COTYLEDONS 1-3 act redundantly to specify the cotyledon boundary. [36−38] HD-GL2 Arabidopsis thaliana meristem L1 layer (ATML1) An early molecular marker for the establishment of both apical-basal and radial patterns during plant embryogenesis. [39] ANTHOCYANINLESS2 (ANL2) anl2 mutant shows aberrant cellular organization. [40] Class I KNOX gene SHOOTMERISTEMLESS (STM) Tanscription factors regulate the architecture of the SAM by maintaining a balance between cell division and differentiation. [41] GRAS SCARECROW (SCR) Regulates the radial organization of the root. [42] AGO proteins ARGONAUTE (AGO) Participate in post-transcriptional gene silencing and influence stem cell fate specification in both plants and animals. [43] PcG proteins POLYCOMB REPRESSIVE COMPLEX
subunit genesEpigenetic effector proteins; stem cell self-renewal, pluripotency, gene silencing; repressive effect on dedifferentiation ability of cells. [15,16] Table 1.
List of some of the major regulatory genes in somatic embryogenesis.
Figures
(1)
Tables
(1)