-
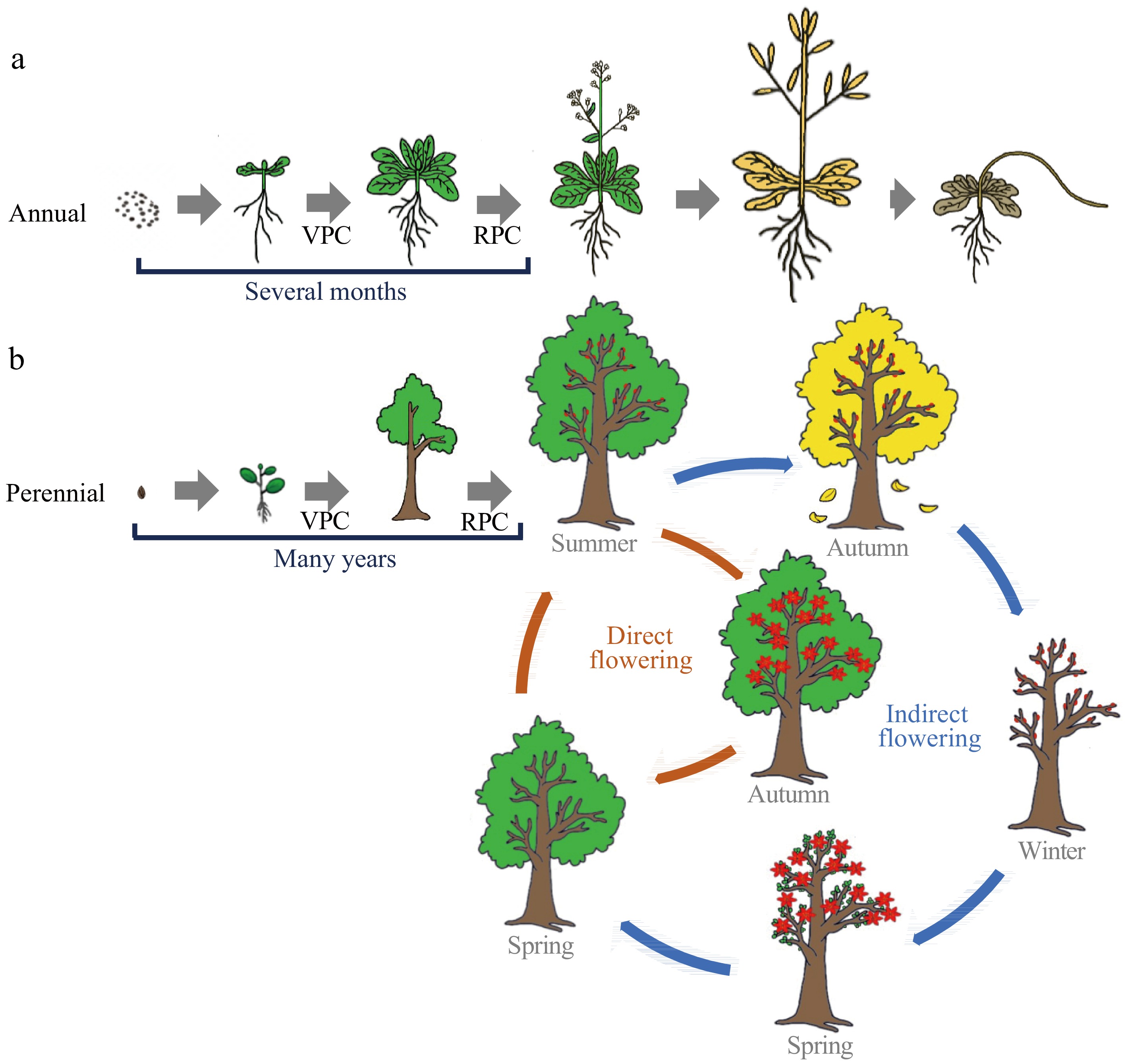
Figure 1.
Comparison of flowering phenology between annual and perennial woody plants. The life cycle of flowering plants can be considered as a succession of distinct growth phases: vegetative growth, followed by a reproductive phase and eventually seed set and senescence. Annuals are fast cyclers and only need several months from the stage of vegetative development to flowering, and complete their life cycle within one growing season (a). While perennial woody plants experience a prolonged vegetative phase with many years until the first onset of flowering. Following first-time flowering, trees undergo seasonal flowering throughout their lifespan (b). Tree’s seasonal flowering can mainly be divided into 'direct' and 'indirect' flowering types, based on whether the development from initiation to emergence is interrupted or includes a period of rest. The 'indirect' flowering is common among temperate/boreal trees. It displays extended periods between flower initiation and flower blooming, in which flowers initiate in the summer are dormant through the winter, and the trees do not blossom until the following spring. In comparison, 'direct' flowering is common among subtropical or tropical evergreen species. They finish their complete reproductive cycles during a single growing season without dormancy or a rest period. VPC, vegetative phase change; RPC, reproductive phase change.
-
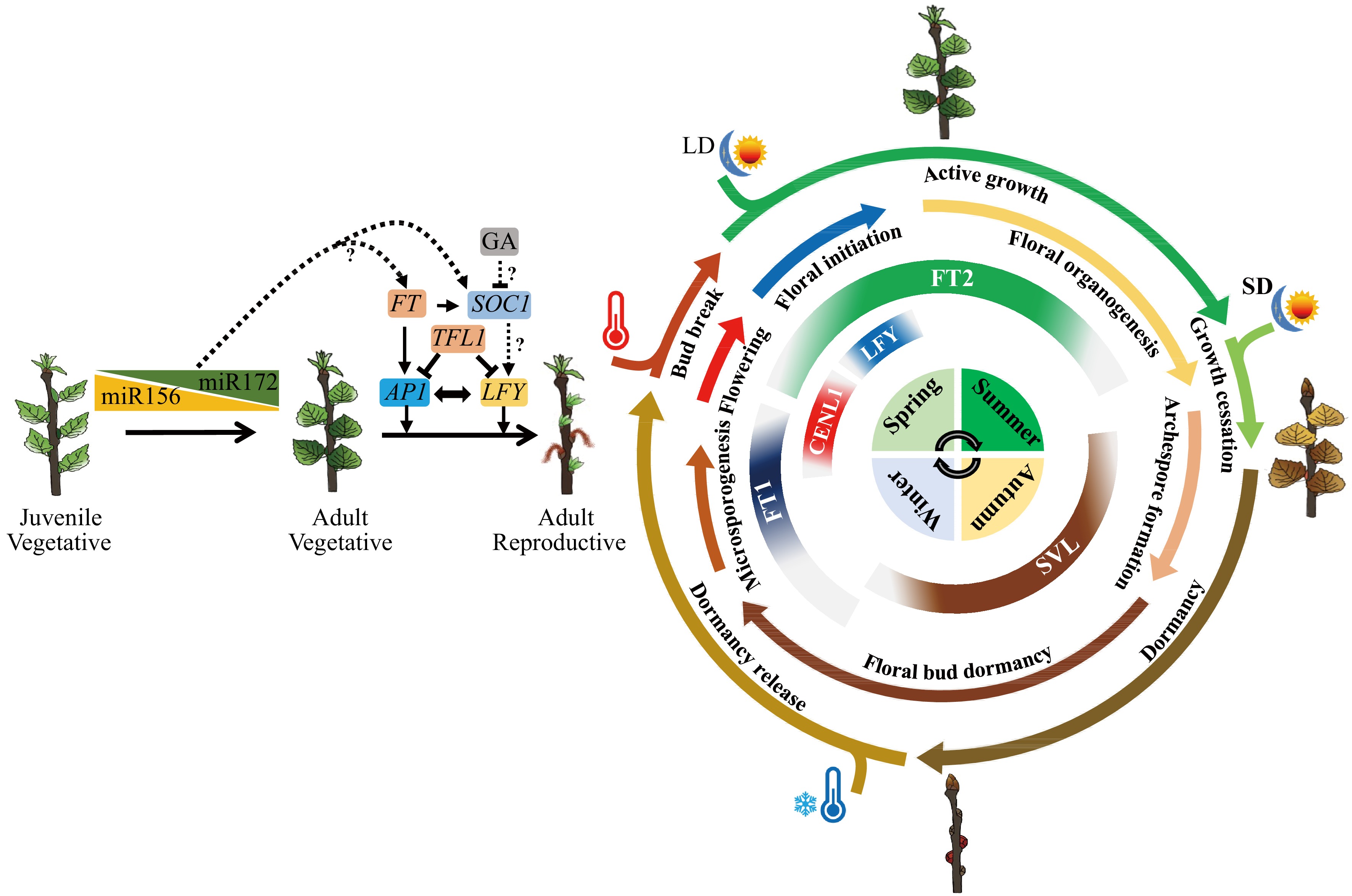
Figure 2.
Molecular pathways of flowering phenology, and their shared mechanisms in seasonal vegetative growth regulation in Populus, the model tree for perennial plant phenology study. Flowering is split into two dimensions: one is the first onset of flowering after many years of juvenile and adult vegetative growth; another is seasonal flowering after reproductive maturity. Conserved to herbaceous plants, the juvenile to adult vegetative phase change is mainly regulated by two microRNAs, miR156 and miR172. The first onset of flowering is controlled by FT/TFL1 family genes and their downstream integrators such as AP1 and LFY. As trees will undergo a long period of adult vegetative stage until floral induction, how miR156/miR172 module regulate age-dependent flowering in trees remains an open question. Unlike in Arabidopsis, GA usually inhibits flowering in diverse woody angiosperms. Whether GA regulates flowering through SOC1-like genes, and do SOC1-like genes control reproductive competence in trees needs further investigation. For seasonal flowering, the expression of flowering integrator genes, such as FT (FT1 and FT2), CENL1, SVL, and LEAFY (LFY), are controlled by seasonal cues like photoperiod and temperature. The specific expression patterns of these genes ensures the tree undergoes floral initiation at a specific time of the year. Meanwhile, these flowering integrator genes also play key roles in the seasonal activity-dormancy vegetative growth, including photoperiod-induced growth cessation of shoot apex at the end of summer, dormancy induction in autumn, cold-induced release of dormancy in winter, and warm temperature-induced bud burst in spring. Thus, trees have evolved an ability to incorporate the environmental signal to different developing events. the diagram sketch of seasonal growth from the inside out represent seasons, the expression pattens of flowering integrator genes, the seasonal flowering events, the seasonal vegetative growth events and environment signals such as photoperiod (LD and SD, long day and short day) and temperature (high and low) respectively.
-
Species Gene Construction Flowering Other effects References Apple
(Malus pumila Mill.)MdFT1 Overexpression A,NInduction [43] MdTFL1, MdTFL1.1
MdCENa, MdCENbCRISPR/RNAi A,T,NRepression [45,159−161] AFL1, AFL2 Overexpression A,NInduction [162,163] MdDAMa, MdDAMb, MdDAMc
MdSVPa, MdSVPbRNAi NInduction Regulates bud dormancy [135] MdFLC1
MdFLC3Overexpression ARepression Juvenility regulation [164] Avocado
(Persea americana)PaFT Overexpression AInduction [165] Blueberry
(Vaccinium corymbosum L.)VcFT Overexpression T,NInduction [166] Birch (Betula) BpAP1 Overexpression NInduction [68] Citrus (Citrus sinensis) CsTFL Overexpression ARepression [167] Citrus
(Citrus clementina)CsAP1 Stress-inducible promoter NInduction [168] CsLFY Stress-inducible promoter NInduction [168] CsSL1, CsSL2 Overexpression AInduction [52] Dogwood (Cornus L.) CorcanTFL1, CorfloTFL1 Overexpression ARepression [169] Eucalyptus
(Eucalyptus spp.)AtFT Overexpression NInduction [152] PtFT1 Overexpression NInduction ELFY CRISPR Affects floral development [170] EgSVP Overexpression ARepression Affects floral development [171] Fig (Ficus carica) FcFT1 Overexpression TInduction [172] Magnoliaceae MawuAP1 Overexpression AInduction [173] Grapevine (Vitis spp.) VvTFL1A Overexpression ARepression [174] VvFT Overexpression AInduction Japanese apricot
(Prunus mume)PmFT Overexpression AInduction [175] PmTFL1 Overexpression ARepression Jatropha
(Jatropha curcas L.)JcFT Overexpression /RNAi A,NInduction [36,176] JcLFY Overexpression A,NInduction Affects floral fruit and seed development [177,178] JcAP1 Overexpression AInduction [179] JcTFL1a, JcTFL1b, JcTFL1c Overexpression A,NRepression [180] JcTFL1 RNAi NInduction Kiwifruit
(Actinidia spp.)AcFT1, AcFT2 Overexpression A,NInduction [181,182] AcCEN1, AcCEN2,
AcCEN3, AcCEN4Overexpression/CRISPR ARepression [47,181,182] AcBFT1, AcBFT2, AcBFT3 Overexpression/CRISPR ARepression Affects dormancy and bud break [182,183] SVP1-4 Overexpression A,T,NNormal Affects dormancy [142,184,185] AcSOC1e, AcSOC1f, AcSOC1i Overexpression AInduction
NNormalAffects dormancy [54] AcFLCL Overexpression/CRISPR Regulate bud break [123] Litchi
(Litchi chinensis Sonn.)LcFT1, LcFT2 Overexpression A,TInduction [186] London plane
(Platanus acerifolia)PaFT Overexpression AInduction
TInduction[187] Longan
(Dimocarpus longan L.)DlFT1 Overexpression AInduction [188] DlFT2 Overexpression ARepression Loquat
(Eriobotrya japonica)EjTFL1-1, EjTFL1-2 Overexpression ARepression [189] EjSOC1-1, EjSOC1-2 Overexpression Acenter [51] EjLFY-1 Overexpression SInduction [190] Mango
(Mangiferaindica L.)MiFT1 Overexpression AInduction [191] MiFT2; MiTFL1-1, MiTFL1-2,
MiTFL1-3, MiTFL1-4Overexpression ARepression [191,192] Norway spruce
(Picea abies)PaFTL1, PaFTL2 Overexpression ARepression [193] PaFTL2 Overexpression Control growth arrest [119] Olive (Olea europaea L.) OeFT1, OeFT2 Overexpression AInduction [194] Peach
(Prunus persica L.)PpTFL1 Overexpression ARepression [195] PpAP1 Overexpression AInduction [196] PpFT Overexpression AInduction [197] Pear
(Pyrus communis L.)PcTFL1-1, PcTFL1-2 RNAi NInduction [151] PcFT2 Overexpression TInduction
NNormalRegulate vegetative growth [198] PcTFL1.1 CRISPR NInduction [159] Pomegranate
(Punica granatum L.)PgTFL1, PgCENa Overexpression ARepression [199] Poplar
(Populus spp.)FT1, FT2 Overexpression/CRISPR NInduction FT1 regulates bud break; FT2 regulates growth cessation [34,44,
103,117]LAP1 Overexpression/RNAi AInduction Regulates growth cessation [200] PopCEN1, PopCEN2 Overexpression/RNAi NRepression Regulates bud break [46] SVL Overexpression/RNAi NRepression Regulate growth cessation, dormancy and bud break [139,140,
143,144]Rubber trees
(Hevea brasiliensis)HbMFT1 Overexpression ARepression [201] Sweet Cherry
(Prunus avium L.)PavFT Overexpression AInduction [202] PavSVP Overexpression ARepression [203] PavSOC1 Overexpression AInduction [55] PaAP1 Overexpression AInduction [204] Tea-oil tree
(Camellia oleifera Abel.)CoFT1 Overexpression AInduction [205] Trifoliate orange
(Poncirus trifoliate)CiFT Overexpression A,NInduction [35,38,206] N, A, T and S represent function assessed in native plant, Arabidopsis, tobacco and strawberry respectively. Table 1.
Functional orthologs of flowering integrator genes identified in perennial trees.
Figures
(2)
Tables
(1)