-
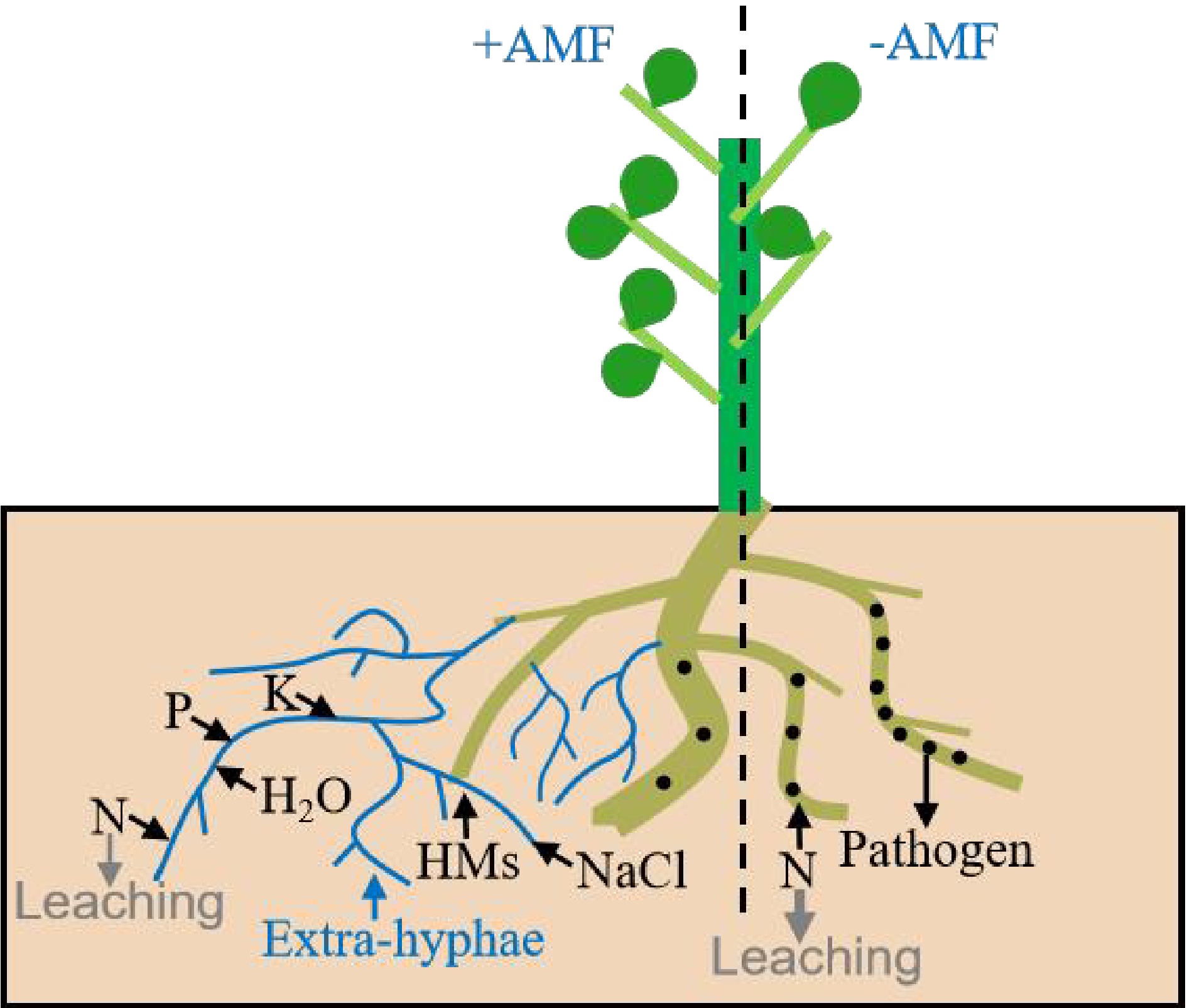
Figure 1.
The formation of AM symbiosis improves the stress tolerance of host plants. The phosphorus (P), nitrogen (N), potassium (K) and other essential nutrients in soil are transferred to the host plants via extraradical hyphae to increase the quality and yield of host plants. The formation of AM symbiosis also enhances the defense of the host plants against various pathogens and the tolerance to HMs (heavy metals) and salt stresses. The thickness of the gray arrow represents the amount of N leaching.
-
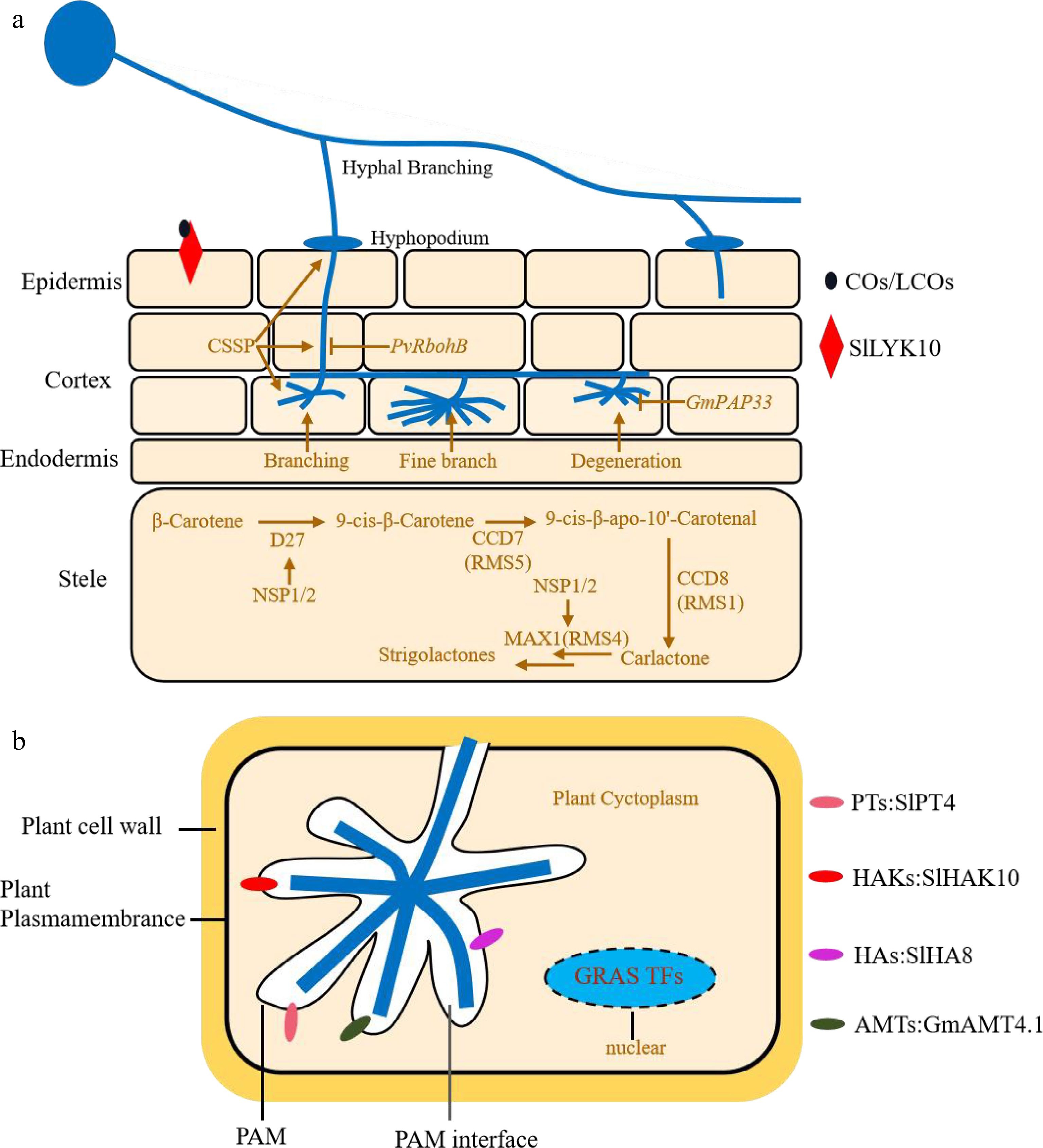
Figure 2.
The molecular mechanism of AM symbiosis in vegetables. (a) RMS1/4/5 are involved in biosynthesis of SLs that promote the spore germination and prime the branching of extraradical hyphae. SlLYK10 binds to COs/LCOs that are secreted by AMF to regulate AM symbiosis. CSSP is required for the early invasion of intra-hyphae and branching of arbuscule. PvRbohB negatively affects the early invasion of intra-hyphae by controlling ROS levels in beans. GmPAP33 negatively regulates the arbuscule degeneration by promoting the hydrolysion of phosphatidylcholine and phosphatidic acid. (b) Many proteins located on PAM, such as SlPT4, SlHA8, SlHAK10 and GmAMT4.1, are required for the exchange of the nutrients and signals between AM and host plants. In addition, GRAS proteins, such as DELLA, RAM1 and RAD1, can regulate the formation of arbuscule.
-
Abbreviated name Full name Number of spore walls Mycorrhizal structures stained
by trypan blueA. laevis Acaulospora laevis 3 V, A, H C. claroideum Claroideoglomus claroideum 1 V, A, H C. etunicatum Claroideoglomus etunicatum 1 V, A, H D. nigra Dentiscutata nigra 3 A, H F. monosporus Funneliformis monosporus 1 V, A, H F. mosseae Funneliformis mosseae 1 V, A, H G. albida Gigaspora albida 1 A, H R. clarum Rhizoglomus clarum 1 V, A, H R. intraradices Rhizoglomus intraradices 1 V, A, H R. irregularis Rhizophagus irregularis 1 V, A, H S. sinuosa Sclerocystis sinuosa 1 V, A, H V, vesicles; A, arbuscules; H, hyphae. -
Gene Organism Gene function Mutant phenotype involved in AM symbiosis Reference RMS1 Pea Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [40] RMS4 Pea Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [41] RMS5 Pea Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [40] NA Pea Required for the biosynthesis of GAs Increased the colonization levels of AM symbiosis [46] SlCCD7 Tomato Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [44] SlIAA7 Tomato Required for the biosynthesis of SLs Reduced the colonization levels of AM symbiosis [45] SlLYK10 Tomato Required for the perception of LCOs Reduced the colonization levels of AM symbiosis [48] SlLYK12 Tomato Required for the perception of LCOs Reduced the colonization levels of AM symbiosis [49] SlCCaMK Tomato Required for the calcium ion spiking in the nuclear Reduced the early invasion of intra-hyphae and the branching of arbuscule [48] SlCYCLOPS Tomato Required for the induction of RAM1 Reduced the early invasion of intra-hyphae and the branching of arbuscule [51] GmSYMRKα/β Soybean Required for the calcium ion spiking in the nuclear Reduced the early invasion of intra-hyphae [52] PvRbohB Bean Required for the production of ROS Increased the early invasion of intra-hyphae [53] CRY/LA
(DELLA)Pea Required for the induction of RAM1 and interacted with MYB1 Inhibited the branching and the degeneration of arbuscule [46] SlDLK2 Tomato Remains unknown Increased the colonization levels of AM symbiosis [60] SlPT4 Tomato Required for the transporting of phosphate Reduced the colonization levels of AM symbiosis [54] SlHA8 Tomato Required for the generation of H+ gradient Inhibited the branching of arbuscule [57] SlHAK10 Tomato Required for the transporting of potassium Reduced the colonization levels of AM symbiosis [56] TSB Tomato Microtubules-associated gene Reduced the colonization levels of AM symbiosis [62] GmPAP33 Soybean Required for the promotion of hydrolysis of phosphatidylcholine and phosphatidic acid Increased the percentage of small arbuscule [63] Table 2.
The genes involved in the formation of AM symbiosis in different crop species.
Figures
(2)
Tables
(2)