-
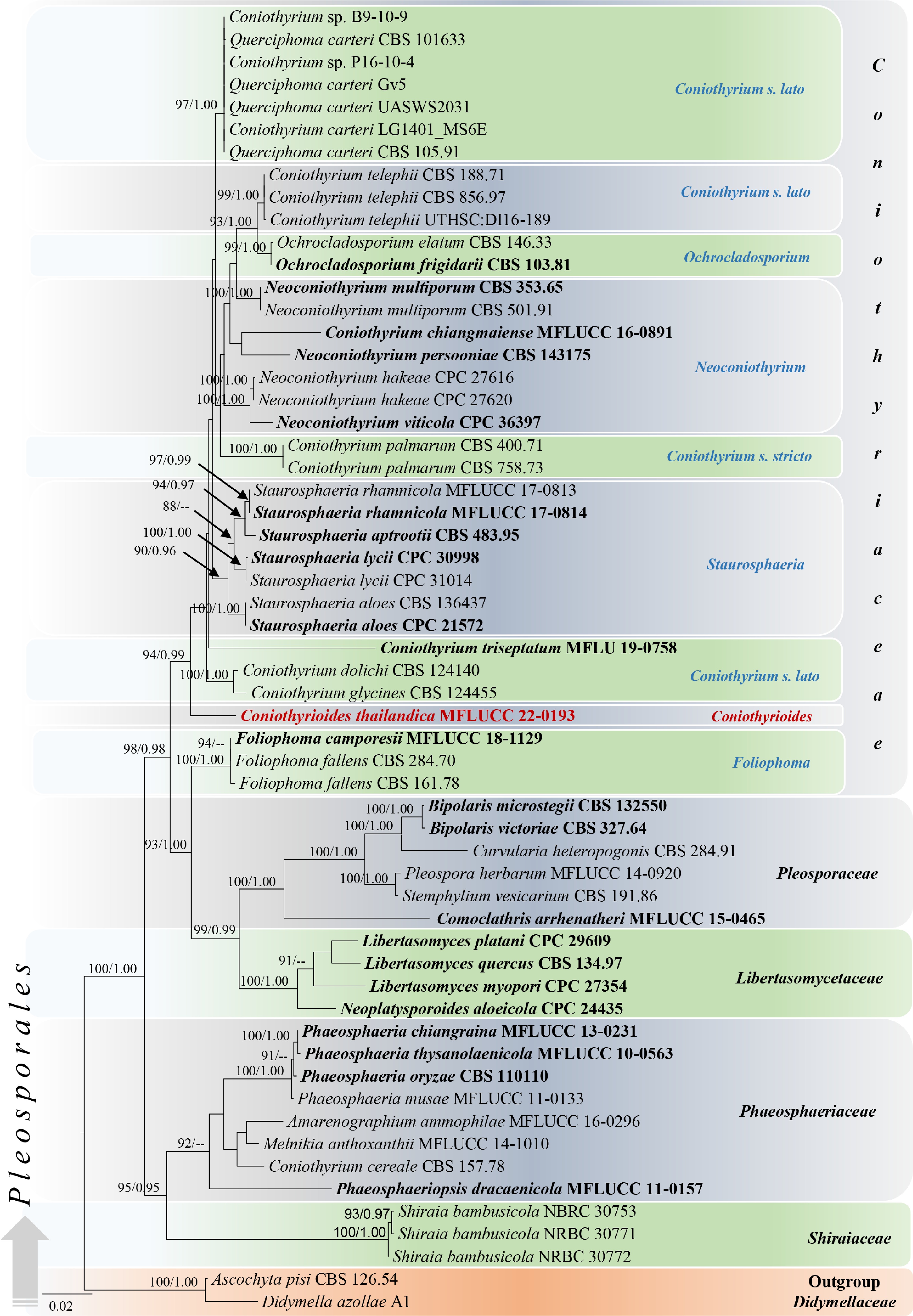
Figure 1.
Phylogram generated from maximum likelihood analysis based on combined LSU, SSU, and ITS sequenced data. Fifty-eight strains were included in the combined sequence analyses, which comprised 2251 characters with gaps (LSU = 800, SSU = 948, ITS = 503). Single gene analyses were also performed, and topology and clade stability were compared from the combined gene analyses. Ascochyta pisi Lib. (CBS 126.54) and Didymella azollae E. Shams, F. Dehghanizadeh, A. Pordel & M. Javan-Nikkhah (A1) were used as the outgroup taxa. The final ML optimization likelihood is -10163.644. The matrix included 494 distinct alignment patterns including undetermined characters. Estimated base frequencies were obtained as follows: A = 0.245, C = 0.219, G = 0.274, T= 0.262; substitution rates AC = 2.73290, AG = 3.93954, AT = 2.73290, CG = 1.0, CT = 7.93321, GT = 1.0 and the gamma distribution shape parameter α = 0.439534. Bootstrap support values for ML (first set) equal to or greater than 75% and BYPP equal to or greater than 0.95 are given above or below the nodes. The strains from the current study are in red bold and the type strains are in black bold. The scale bar represents the expected number of nucleotide substitutions per site.
-

Figure 2.
Coniothyrioides thailandica sp. nov. (MFLU 22-0276, holotype). (a) & (b) Appearance of conidiomata on a submerged decaying woody substrate. (c) Longitudinal section of conidioma. (d) Conidiomatal wall. (e) The appearance of setae. (f) & (g) Conidiogenous cells with developing conidia. (h) Conidia. Scale bars: a = 200 μm, b = 100 μm, c = 50 μm, d = 20 μm, e, h = 10 μm, f, g = 5 μm.
-
Genes/loci PCR primers (forward/reverse) PCR conditions Reference (s) ITS and LSU ITS5/ITS4 and LR0R/LR5 94 °C; 2 min (95 °C; 30 s, 55 °C; 50 s, 72 °C; 90 s) × 35 thermal cycles, 72 °C; 10 min [ 42− 44] SSU NS1/NS4 95 °C; 3 min (95 °C; 30 s, 55 °C; 50 s, 72 °C; 30 s) × 35 thermal cycles, 72 °C; 10 min [ 42] Table 1.
Gene regions, primers, and PCR thermal cycle programs used in this study, with corresponding reference(s).
-
Species Strain/voucher number GenBank accession numbers ITS LSU SSU Amarenographium ammophilae MFLUCC 16–0296 KU848196 KU848197 KU848198 Ascochyta pisi CBS 126.54 GU237772 EU754137 EU754038 Bipolaris microstegii CBS 132550 NR_120160 NG_042690 NA Bipolaris victoriae CBS 327.64 NR_147489 NG_069233 NA Comoclathris arrhenatheri MFLUCC 15–0465 NR_165855 NG_068240 NG_068374 Coniothyrioides thailandica MFLUCC 22-0193 OQ023276 OQ023277 OQ025050 Coniothyrium carteri LG1401 MS6E KX359604 KX359604 NA Coniothyrium cereale CBS 157.78 MH861123 JX681080 NA Coniothyrium chiangmaiense MFLUCC 16–0891 KY568987 KY550384 KY550385 Coniothyrium dolichi CBS 124140 JF740183 GQ387611 GQ387550 Coniothyrium glycines CBS 124455 JF740184 GQ387597 GQ387536 Coniothyrium palmarum CBS 400.71 AY720708 EU754153 AY720712 Coniothyrium palmarum CBS 758.73 NA JX681085 EU754055 Coniothyrium sp. B9-10-9 MW764153 NA NA Coniothyrium sp. P16-10-4 MW764259 NA NA Coniothyrium telephii CBS 188.71 JF740188 GQ387599 GQ387538 Coniothyrium telephii CBS 856.97 JF740189 GQ387600 GQ387539 Coniothyrium telephii UTHSC:DI16–189 LT796830 LN907332 NA Coniothyrium triseptatum MFLU 19–0758 NR_171948 NG_073674 NA Curvularia heteropogonis CBS 284.91 JN192379 JN600990 NA Didymella azollae A1 MT514913 MT514910 NA Foliophoma camporesii MFLUCC 18–1129 KY929151 KY929181 NA Foliophoma fallens CBS 161.78 KY929147 GU238074 GU238215 Foliophoma fallens CBS 284.70 KY929148 GU238078 GU238218 Libertasomyces myopori CPC 27354 NR_145200 NG_058241 NA Libertasomyces platani CPC 29609 NR_155336 NG_059744 NA Libertasomyces quercus CBS 134.97 NR_155337 DQ377883 NA Melnikia anthoxanthii MFLUCC 14–1010 NA KU848204 KU848205 Neoconiothyrium hakeae CPC 27616 KY173397 KY173490 NA Neoconiothyrium hakeae CPC 27620 KY173398 KY173491 NA Neoconiothyrium multiporum CBS 353.65 NR_111617 JF740268 NA Neoconiothyrium multiporum CBS 501.91 JF740186 GU238109 GU238225 Neoconiothyrium persooniae CBS 143175 NR_156386 NG_058509 NA Neoconiothyrium viticola CPC 36397 NR_165929 NG_068326 NA Neoplatysporoides aloeicola CPC 24435 NR_154230 NG_058160 NA Ochrocladosporium elatum CBS 146.33 EU040233 EU040233 NA Ochrocladosporium frigidarii CBS 103.81 NR_156512 NG_064123 NA Phaeosphaeria chiangraina MFLUCC 13–0231 KM434270 KM434280 KM434289 Phaeosphaeria musae MFLUCC 11–0133 KM434267 KM434277 KM434287 Phaeosphaeria thysanolaenicola MFLUCC 10–0563 NR_155642 NG_069236 NG_063559 Phaeosphaeria oryzae CBS 110110 NR_156557 NG_069025 NG_061080 Phaeosphaeriopsis dracaenicola MFLUCC 11–0157 NR_155644 NG_059532 KM434292 Pleospora herbarum MFLUCC 14-0920 KY659560 KY659563 KY659567 Querciphoma carteri CBS 101633 JF740180 GQ387593 GQ387532 Querciphoma carteri CBS 105.91 JF740181 GQ387594 GQ387533 Querciphoma carteri Gv5 MT819903 NA NA Querciphoma carteri UASWS2031 MN833930 NA NA Shiraia bambusicola NBRC 30753 AB354987 AB354968 NA Shiraia bambusicola NRBC 30771 AB354990 AB354971 NA Shiraia bambusicola NRBC 30772 AB354991 AB354972 NA Staurosphaeria aloes CBS 136437 KF777142 KF777198 NA Staurosphaeria aloes CPC 21572 NR_137821 NG_067283 NA Staurosphaeria aptrootii CBS 483.95 NR_155186 NA NA Staurosphaeria lycii CPC 30998 KY929150 KY929180 NA Staurosphaeria lycii CPC 31014 KY929151 KY929181 NA Staurosphaeria rhamnicola MFLUCC 17–0813 MF434200 MF434288 MF434376 Staurosphaeria rhamnicola MFLUCC 17–0814 NR_154461 MF434289 NG_063659 Stemphylium vesicarium CBS 191.86 MH861935 MH873624 GU238232 NA: Sequences not available in GenBank. Table 2.
Taxa used in the phylogenetic analyses and their GenBank accession numbers. Sequences of new taxon generated in this study are in blue-bold and type strains are in black-bold.
-
Species Conidiomata (µm) Conidiomata wall (µm) Conidiogenous cells (µm) Conidia (µm) Habitat(s) and host(s) Locality Reference Coniothyrioides thailandica (holotype: MFLU 22-0276) 150–200 high, × 100–150 diam., pycnidial, semi-immersed, erumpent, dark brown to black, globose to subglobose,
uni-locular, ostiolate15–20 wide, black, dark brown to hyaline cells of textura angularis,
Brown, septate setae (3–5 µm wide,) with hyaline apex4–5 long × 2.5–3.5 wide, hyaline, doliiform to subcylindrical, enteroblastic, phialidic conidiogenesis with periclinal thickening 3–5 × 2.5–3, ellipsoidal to obovoid, aseptate, rounded at apex, initially hyaline, becoming pale to dark brown at maturity On decaying wood in salt marsh habitat Thailand This study Coniothyrium
palmarum (CBS 400-71)Immersed, dark brown, globose, pale to uni-locular brown, thick-walled cells of textura angularis hyaline, phialidic conidiogenesis, doliiform to cylindrical Subcylindrical, spherical, ellipsoid or broadly clavate, 0(–1)-septate, apex obtuse, brown, base truncate, sometimes minute marginal frill On Chamaerops humilis ( Arecaceae) Italy [ 16]PP,
[ 20]GNFoliophoma fallens (holotype: CBS 284.70) 120–250 wide, eustromatic, globose, uni-multi locular,
1–3 ostiolate3–6 layers,
brown textura angularis5–7 × 4–5, hyaline, phialidic conidiogenesis with thickening or proliferation at apex, dolliform to subcylindrical,
periclinal(5–)5.5–6(–7) × (3–)4(–5),
broadly ellipsoidal, aseptate, hyaline, guttulate or granular, apex obtuse, base truncate to bluntly roundedLeaf spot on Nerium oleander ( Apocynaceae) Italy [ 27] *Foliophoma camporesii (holotype: MFLU 17-1006) 40–47 × 40–69, pycnidial, immersed to semi-immersed, globose to subglobose, ellipsoidal or irregular, carbonaceous 15–40, 1–2-layered of cells of textura angularis 2–4 × 2–3, hyaline, globose to short cylindrical, phialidic conidiogenesis with periclinal thickening or percurrent proliferation at apex 2–6 × 3–5, ovoid to ellipsoidal, aseptate, hyaline when immature, brown at maturity On dead stems of Maclura pomifera ( Moraceae) Italy [ 25] Hazslinszkyomyces aloes (≡ Camarosporium aloes: ex-type - CPS 21572) 250 diam, pycnidial, erumpent, brown, globose, central ostiolate 3–6 layers of brown textura angularis
5–10 × 4–5, hyaline,
ampulliform to doliiform, apex with several inconspicuous percurrent proliferation,(9–)11–13(–14) × (4–)6–7(–8), ellipsoid, initially hyaline, aseptate, becoming pale brown, subcylindrical
to clavate or obovoid with 3 transverse eusepta, constricted at median septum or not, apex obtuse, base bluntly rounded to truncateDead bark of Aloe dichotoma ( Xanthorrhoeaceae) South Africa [ 27] Neoconiothyrium persooniae (ex-type CPC 32021 = CBS 143175) 100–200 diam, superficial, ellipsoid to obpyriform, 1–2 papillate ostioles, 10–15 diam, with or without setae 3–6 layers, hyaline textura angularis 5–8 × 4–5, hyaline, doliiform to ampulliform, phialidic, with periclinal thickening or percurrent proliferation (5–)6–7(–8) × 3(–4), ellipsoid to subclavate, aseptate, initially hyaline medium brown, becoming cylindrical and at times 1-septate, apex subobtuse, base bluntly rounded On leaves of Persoonia laurina subsp. laurina ( Proteaceae) Australia [ 24] Ochrocladosporium elatum (CBS 146.33) – – Integrated as lateral peg-like loci on hyphal
cells, or erect, subcylindrical, up to 25 µm long, 2.5–4 µm wide,
with 1–3 terminal loci, occasionally lateral, 1–1.5 µm wideRamoconidia, 10–40 × 3–5, subcylindrical to ellipsoid, hyaline to pale brown, 0(–1)-septate, giving rise to branched
chains of conidia that are subcylindrical to ellipsoid, aseptate, (7–)8–10(–14) × (3–)4(–4.5), olivaceous brownWood pulp Sweden [ 35] '–' observed morphologies on cultures, therefore conidiomata and wall characters are not recorded. '*' species which is not represent a generic type. GN- based on the generic description. PP- based on the photographic plate provided. Table 3.
Synopsis of asexual morphological characters of related genera of Coniothyriaceae.
-
Species Strain LSU SSU ITS Coniothyrium palmarum CBS 400-71 14/800 (1.75%) 3/948 (0.3%) 69/487 (14.10%) Foliophoma fallens CBS 284.70 8/800 (1%) 2/948 (0.2%) 66/497 (13.27%) Hazslinszkyomyces aloes CPC:21572 6/800 (0.75%) – 52/497 (10.46%) Neoconiothyrium persooniae CBS:143175 20/800 (2.5%) – 49/497 (9.85%) Ochrocladosporium elatum CBS 146.33 13/800 (1.62%) – 53/497 (10.66%) Table 4.
The base pair comparisons of our strain (MFLUCC 22-0193) with the strains representing type species of other genera in Coniothyriaceae.
Figures
(2)
Tables
(4)