-
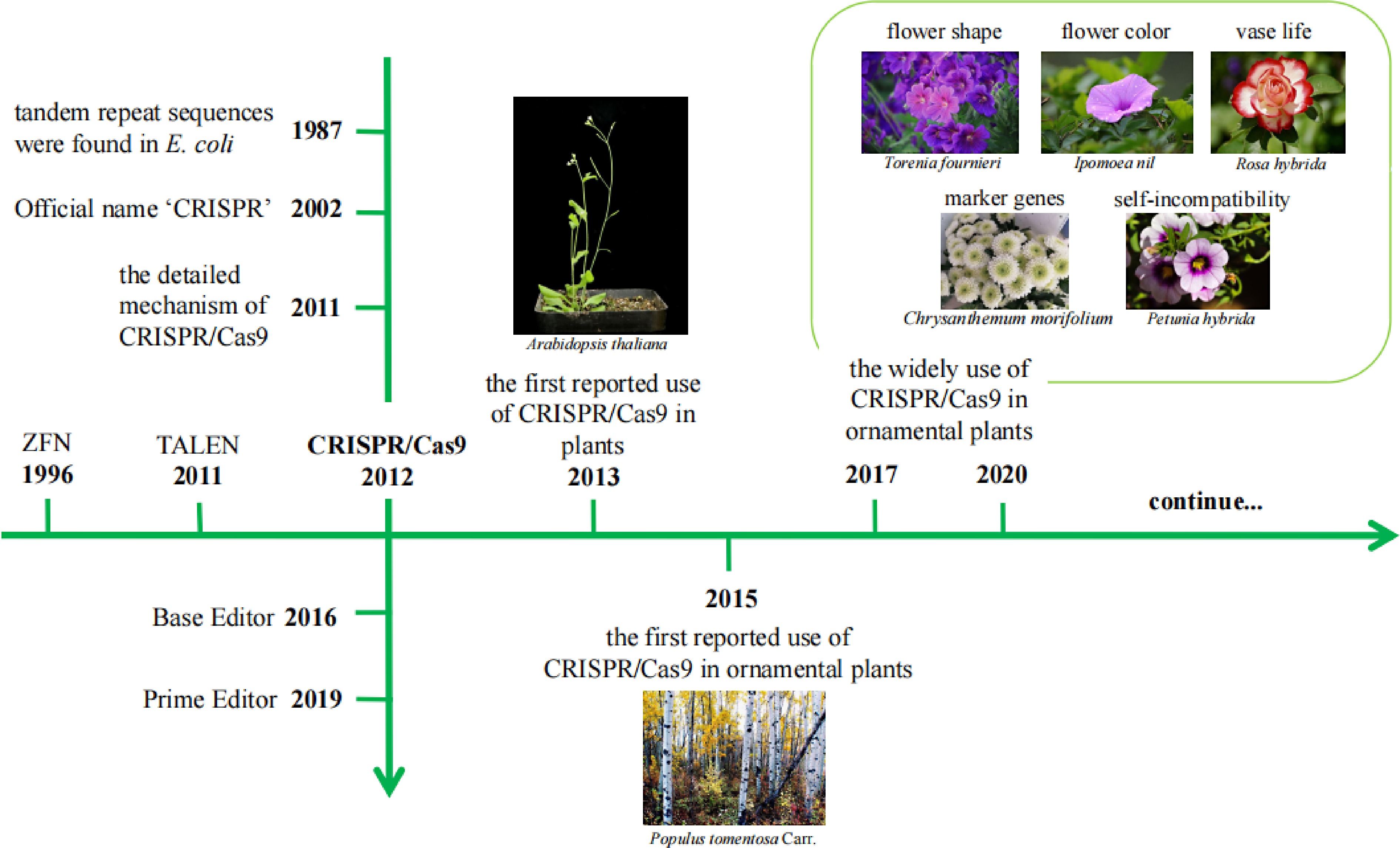
Figure 1.
The timeline of CRISPR/Cas9 development and use in ornamental plants.
-
Plant species Target trait Target gene Material Method Phenotype References Torenia fournieri Flower type RAD1 Agrobacterium-mediated
transformationA violet pigment pattern on dorsal petals [23] Phalaenopsis equestris Flower type MADS Explant Agrobacterium-mediated
transformationInfluence floral organ initiation and development [25] Torenia fournieri Color F3H Leaf Agrobacterium-mediated
transformationFaint blue (almost white) and pale violet flowers [26] Petunia Color F3H Protoplast PEG-mediated
transformationPale purplish pink flower [27] Petunia Color CCD4 Embryo-derived secondary embryo Agrobacterium-mediated
transformationPale yellow petals [28] Japanese Gentian Color GST1 Leaf Agrobacterium-mediated
transformationAlmost white or pale blue [29] Ipomoea nil Color DFR-B Embryo Agrobacterium-mediated
transformationGreen stems and white flowers [30] Ipomoea nil Vase life EPH1 Embryo Agrobacterium-mediated
transformationPetal senescence delay [31] Rosa hybrida Vase life EIN2 Somatic embryos Agrobacterium-mediated
transformationFlower opening completely blocked [32] Petunia hybrida Vase life ACO1 Protoplast PEG-mediated transformation Increased flower vase life [33] Chrysanthemum Marker gene YGFP Young leaf Agrobacterium-mediated
transformationGFP fluorescence [34] Lilium
Longiflorum & Lilium pumilumMarker gene PDS Embryogenic
calli and scalesAgrobacterium-mediated
transformationAlbino, pale yellow and albino green [35] Dendrobium officinale lignin synthesis C3H, C4H, 4CL,
CCR, IRXProtocorm Agrobacterium-mediated
transformationAffected the lignin biosynthesis [36] Petunia inflata reproduction SSK1 Leaves Agrobacterium-mediated
transformationInhibit the growth of pollen tubes [37] Table 1.
List of current research of gene editing in ornamental plants.
Figures
(1)
Tables
(1)