-
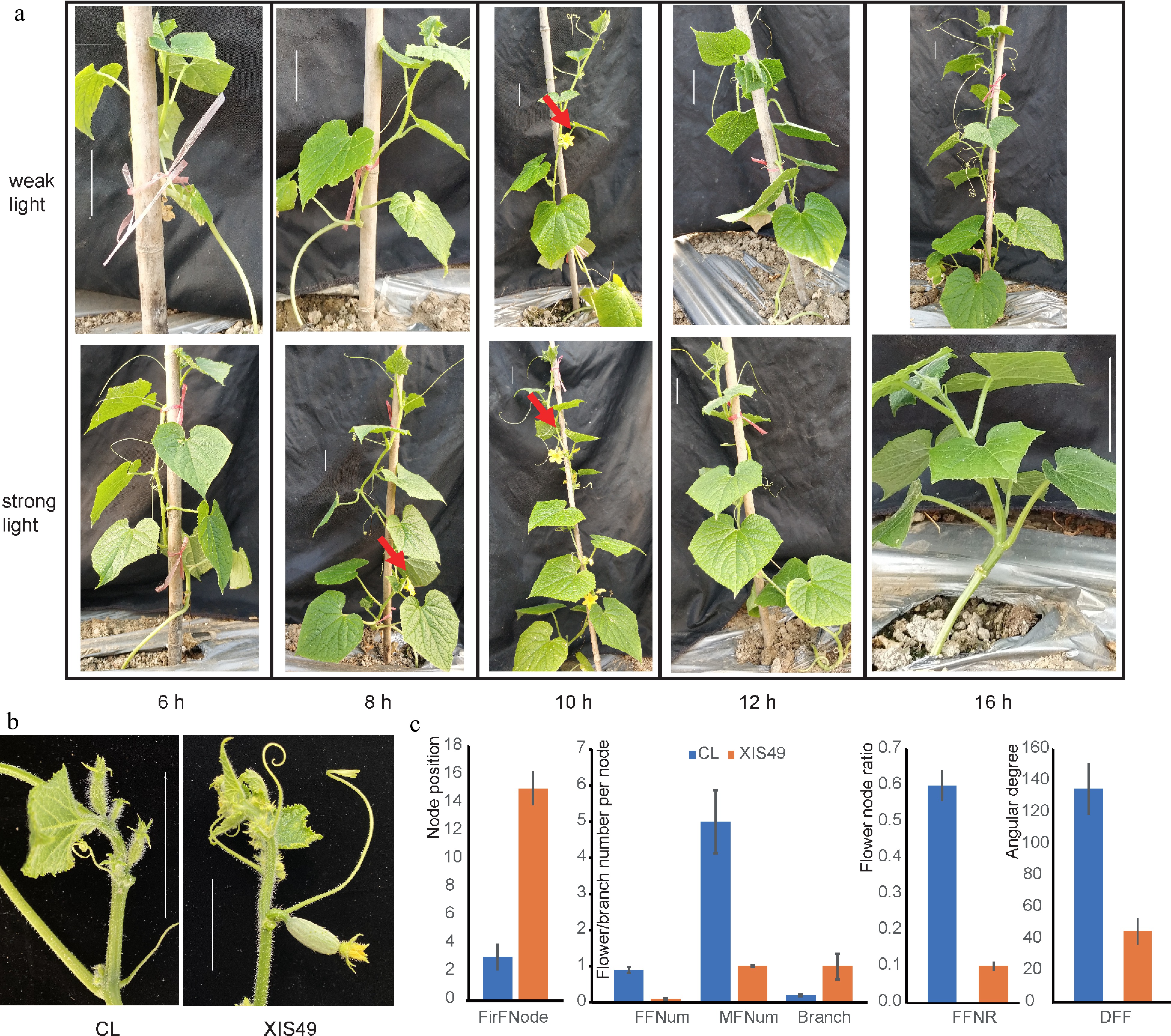
Figure 1.
Characteristic flowering-related phenotypes of XIS49. (a) Photoperiod and light intensity-dependent flowering initiation. The bars indicate a scale of 5 cm. (b) Female flower orientation. The bars indicate a scale of 5 cm. (c) XIS49 is significantly different with CL cucumber in first flower node (FirFNode), female flower number (FFNum) per node, male flower number (MFNum) per node, branch number per node, female flower node ratio (FFNR), and downward female flower (DFF) denoted by the angle at the stem.
-
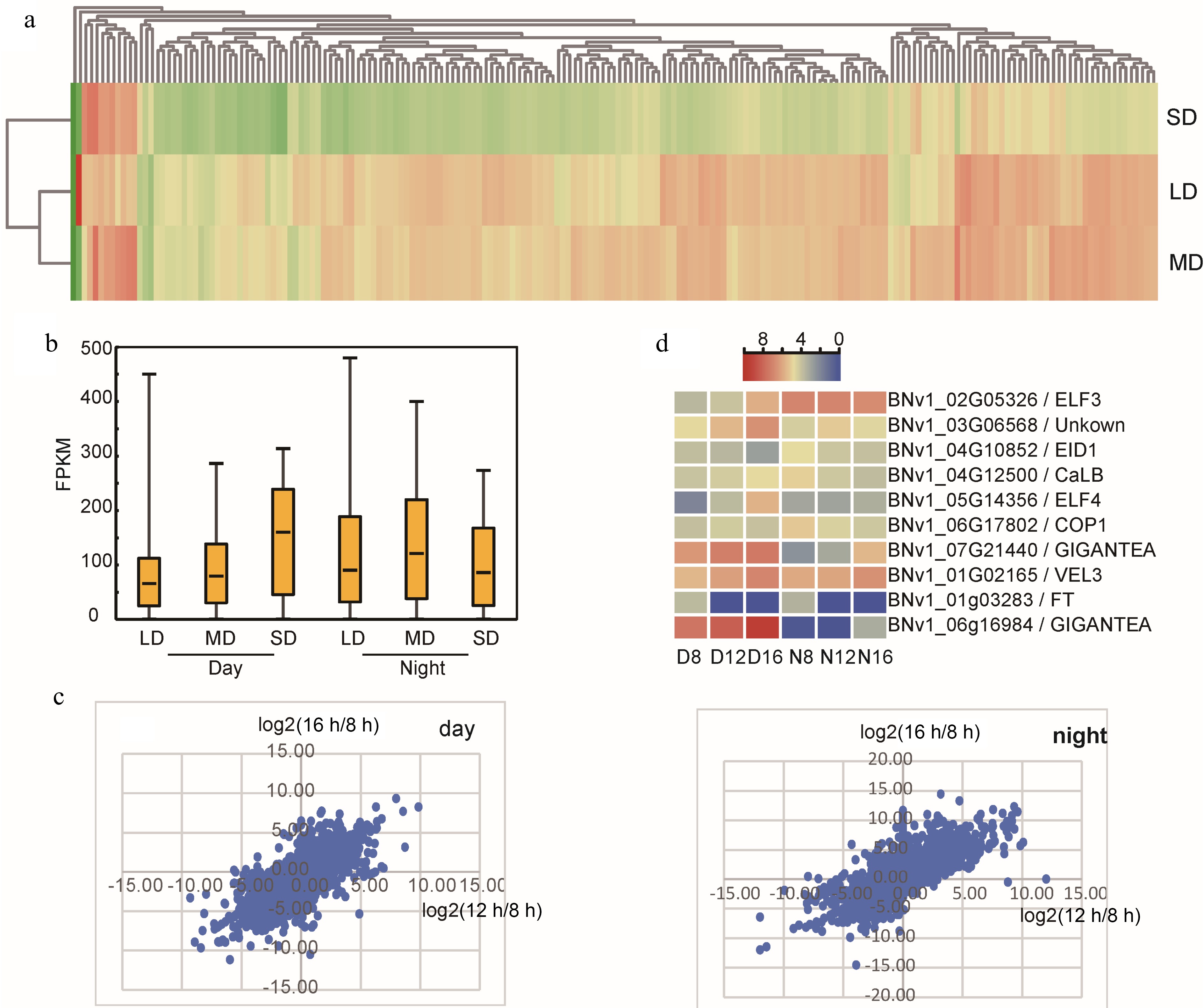
Figure 2.
Influence of photoperiod treatment on XIS49 transcriptome. (a) A great number of ribosome genes show diurnal rhythm, and these day-night ribosome DEGs (day vs night) are upregulated at night under LD and MD but downregulated at night under SD. (b) Photoperiodic DEGs (LD vs SD, MD vs SD, LD vs MD) show much higher expression level under SD in daytime but not in nighttime. (c) Plotting the value of log2(16 h-FPKM/8 h-FPKM) against log2(12 h-FPKM/8 h-FPKM) by using the photoperiodic DEG set. (d) Flowering-related photoperiodic DEGs annotated by GO term.
-
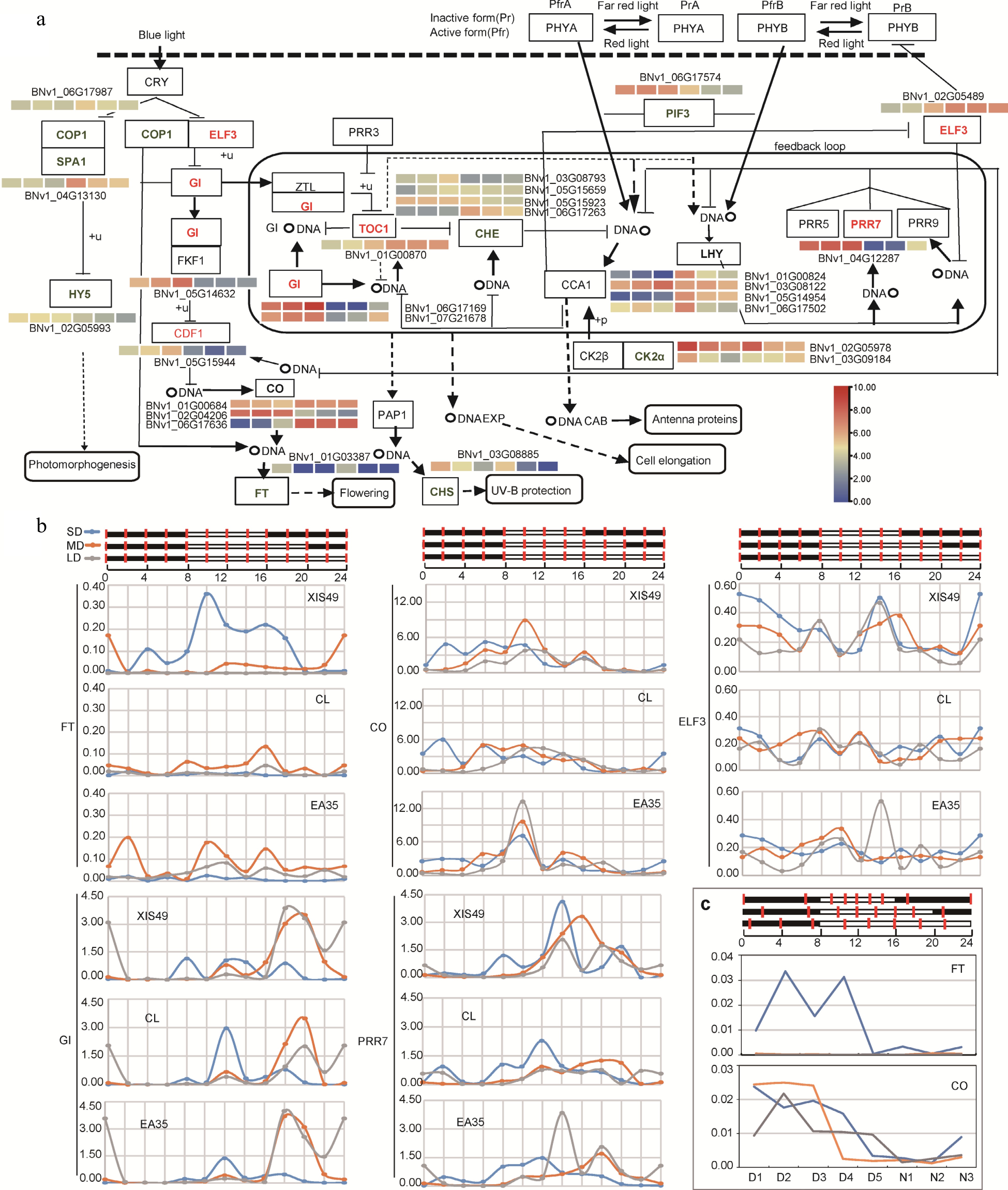
Figure 3.
Circadian expression manner of DEGs (a) Influence of photoperiod treatment on circadian pathway genes (KEGG, ko04712). Upregulated and downregulated genes by long photoperiod is denoted in red and green. A heat map is present to show transcriptional change; from left to right, 8 h-day, 12 h-day, 16 h-day, 8 h-night, 12 h-night, 16 h-night. (b) Samples were collected every two hours during a 24 h period and the expression was quantified by qRT-PCR. (c) The circadian expression of CO and FT was confirmed again by using leaf samples collected at nighttime-trisection timepoints (N1-N5) and daytime-quinquesection timepoints (D1-D5).
-
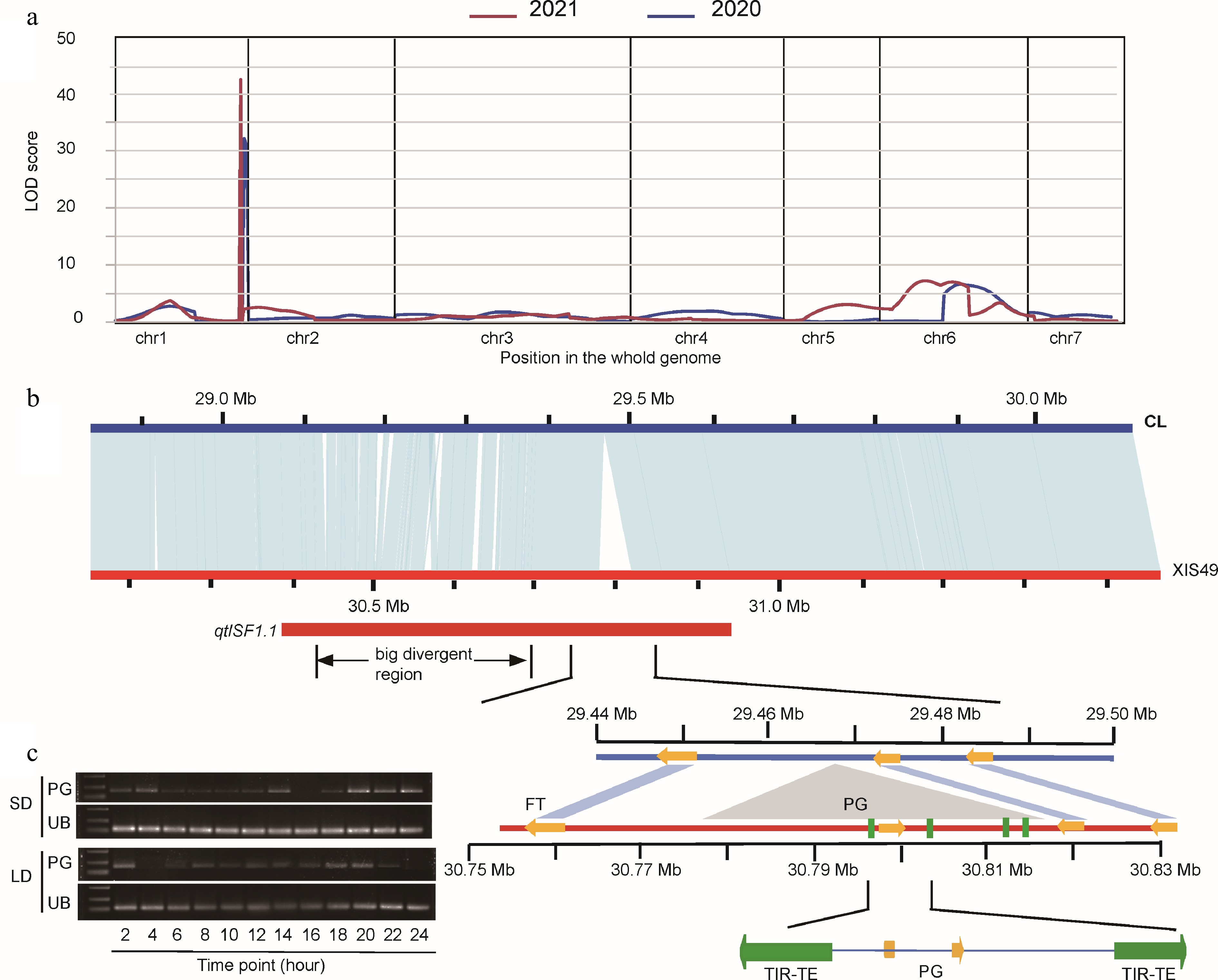
Figure 4.
QTL mapping of short-day flowering in XIS49. (a) LOD profiles of QTL for FFNode (first flower node). Plants were pretreated by LD condition. (b) A syntenic analysis of 1 Mb segment that carries the QTL between XIS49 and CL cucumber. The qtlSF1.1 carry two major sequence variants nearing the FT gene, 'the bid divergent region' and 'the big insertion'. The orange arrows indicate protein coding genes. The four green rectangles indicate TIR-TE insertion. (c) The expression profile in a 24-h period of the inserted polygalacturonase (PG) gene shows photoperiodic manner. UB, ubiquitin.
-
Photoperiod First flower node1 First FF node1 FF number1 8H 3.5 ± 0.2 e 8.9 ± 0.3 c 1.8 ± 0.2 a 10H 4.7 ± 0.3 d 11.3 ± 0.3 b 2.1 ± 0.3 a 10.5H 6.0 ±0.4 c 12.6 ± 0.3 ab 1.5 ± 0.4 a 11H 5.2 ± 0.2 cd 12.9 ± 0.8 a 1.6 ± 0.2 a 11.5H 10.4 ± 0.4 b − − 12H 17.6 ± 0.6 a − − 16H − − − 1 Average flower number ± SE within 25 nodes. The different letters following the numbers indicate significant difference between different photoperiod, P ≤ 0.05. FF, female flower; "−" indicates no flowers observed. Table 1.
Influence of photoperiod on flowering of XIS cucumber.
-
Chr. Start End LOD PVE% Size (Mb) SF1.1 1 29,795,488 29,931,423 32.09 46.90 0.14 SF6.1 6 12,580,505 21,116,696 6.65 7.90 8.54 FirFFNode1.1 1 190,063 10,053,157 8.97 15.72 9.86 FFNum1.1 1 190,063 10,053,157 6.03 8.70 9.86 FFNum2.1 2 12,405,113 22,000,804 5.81 11.55 9.60 MFNum1.1 1 29,369,853 29,795,488 18.10 14.11 0.43 MFNum6.1 6 2,507,906 12,580,505 14.14 16.15 10.07 MFNum6.2 6 12,580,505 21,116,696 14.85 12.18 8.54 FFNR1.1 1 190,063 10,053,157 7.89 13.20 9.86 DFF3.1 3 135,870 11,923,221 6.76 11.00 11.79 branch1.1 1 29,795,488 29,931,423 5.19 7.17 0.14 Short-day flowering (SF) was mapped based on the phenotype of first flower node; FirFFNode, first female flower node; FFNum, female flower number; MFNum, male flower number; FFNR, female flower node ratio; DFF, downward female flower; branch, branch number per node. Table 2.
QTL mapping of several flower traits by KASP.
-
Gene ID Position/Mb SVs in promoter SNP Gene function CDS Promoter2 CsaV3_1G043880 29.04 HDR Unknown CsaV3_1G043890 29.04 DEL; DEL T(-31)A; C(-30)G; A(-5)C Unknown CsaV3_1G043980 29.14 INS; INS T18I; D214N; I304Y Choline monooxygenase CsaV3_1G043990 29.15 HDR A51T A(-26)C Unknown CsaV3_1G044000 29.15 INS Y158S Phosphatidylglycerol transfer protein CsaV3_1G044010 29.17 DEL Unknown CsaV3_1G044020 29.18 CPL DnaJ protein CsaV3_1G044060 29.23 HDR; INS L23H; K127R; A130S; S202L T(-43)A Cytochrome P450 CsaV3_1G044110 29.33 HDR YTH domain, RNA binding CsaV3_1G044160 29.38 HDR Thioredoxin F-type CsaV3_1G044440 29.78 CPG; INS A27T Protein TIC 20-II CsaV3_1G043900 29.05 DEL Unknown CsaV3_1G043920 29.06 DEL T(-43)G; A(-22)T DnaJ protein CsaV3_1G043930 29.08 DEL A572T Serine/threonine-protein phosphatase CsaV3_1G043960 29.12 DEL Glycosyl transferase CsaV3_1G044070 29.25 DEL S locus-related glycoprotein 1 binding pollen coat protein CsaV3_1G044130 29.36 INS Glycoside hydrolase CsaV3_1G044170 29.39 DEL Unknown CsaV3_1G044180 29.43 DEL Activator of Hsp90 ATPase CsaV3_1G043940 29.10 F281I; Q332R; I441T; I574L Unknown CsaV3_1G043950 29.11 E13D; K34E; P26L; Y29F; L67S Unknown CsaV3_1G043970 29.13 Protein IQ-DOMAIN CsaV3_1G044030 29.19 D47N; C81R; V113A; A119G; H145Y; Q160K; F207L; Y211N 23 kDa jasmonate-induced protein CsaV3_1G044040 29.20 F208L; G381D; I424M; S753F; G790D beta-galactosidase 7 CsaV3_1G044050 29.22 G431R; L569S Microtubule-associated protein CsaV3_1G044080 29.28 S20A Pentatricopeptide repeat-containing protein tubulin-folding cofactor E CsaV3_1G044090 29.28 CsaV3_1G044100 29.32 Heavy metal-associated domain CsaV3_1G044120 29.35 S90A Unknown CsaV3_1G044140 29.37 D317E Aspartic proteinase CsaV3_1G044150 29.37 Unknown CsaV3_1G044210 29.45 G82R FT CsaV3_1G044450 29.80 R64G Phosphoglycolate phosphatase 1 Sequential/Structural variance (SV) including (HDR), (CPL), (CPG), and insertion (INS) and deletion (DEL); Regular INS and DEL, SV length > 50 bp; italic INS and DEL, SV length < 50. 2 T(-31)A, the nucleotide with a distance of upstream-31 bp to the transcription start site is T in CL, and A in XIS49. Table 3.
Genes at the locus LF1.1 with sequential divergent between CL and XIS49.
Figures
(4)
Tables
(3)