-
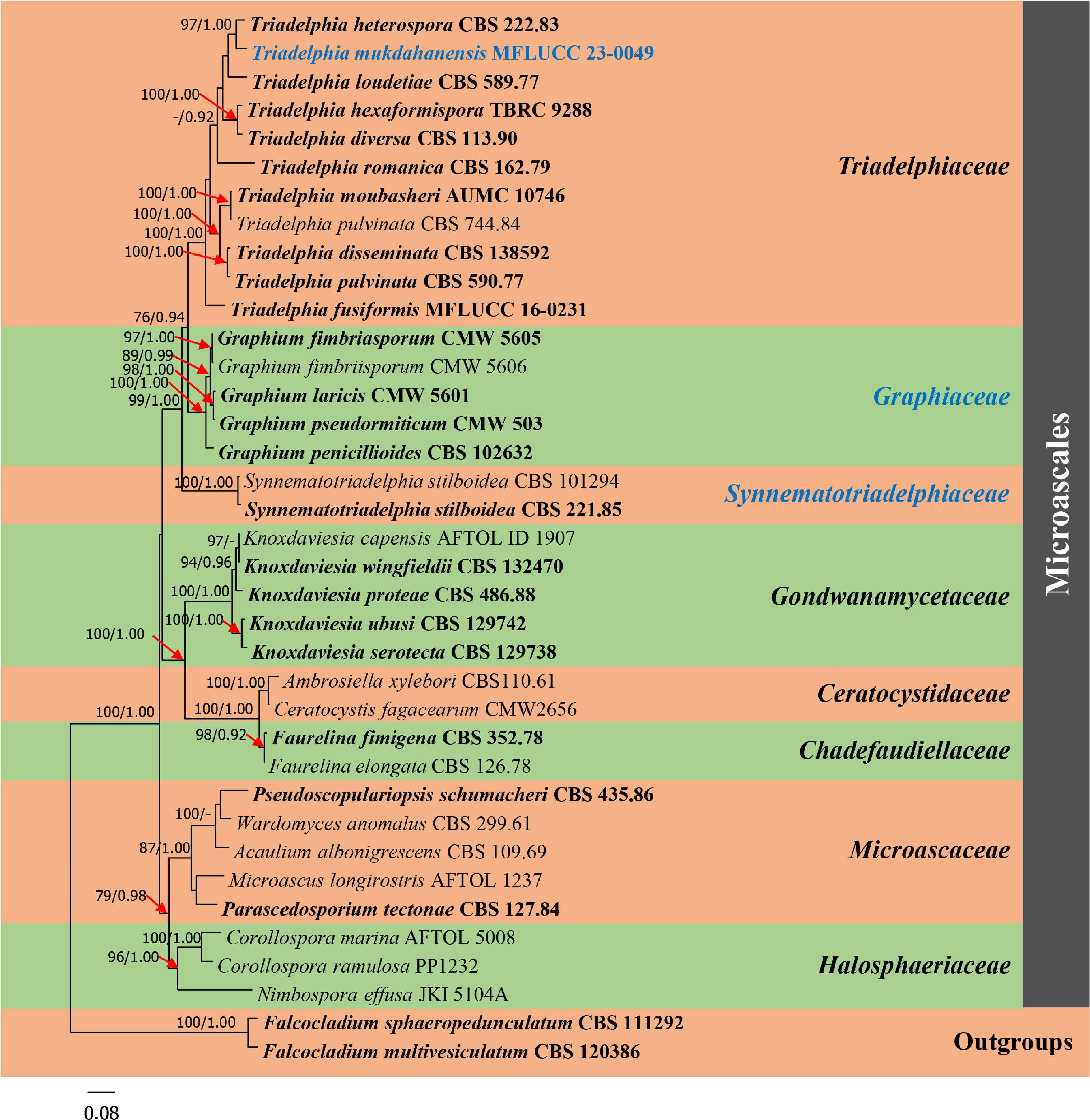
Figure 1.
Phylogram generated from maximum likelihood analysis based on combined dataset of ITS, LSU, SSU and RPB2 sequence data of Microascales group. Bootstrap support values for maximum likelihood (ML) equal to or greater than 75% and Bayesian posterior probabilities (PP) equal to or greater than 0.90 are given above the nodes. A newly generated sequence and a newly introduced family are in blue bold. Type species are in bold. The tree is rooted with Falcocladium sphaeropedunculatum (CBS 111292) and F. multivesiculatum (CBS 120386).
-

Figure 2.
Triadelphia mukdahanensis (holotype). (a)–(d) Appearance of colonies on substrate. (e)–(g) Conidiogenous cells and conidia (e and f mounted in 5% KOH). (f)–(k) conidial type (a). (l)–(n) Conidial type (b). (o)–(p) Conidial type (f). (q) Conidial type (a), (b) and (f). Scale bars: a–c = 500 µm, d = 20 µm, e–i, k–n, q = 10 µm. j, o, p = 5 µm.
-
Taxa Strain no.1 GenBank accession numbers2 ITS LSU SSU RPB2 Acaulium albonigrescens CBS 109.69 KY852469 KY852480 − − Ambrosiella xylebori CBS 110.61 − EU984294 AY858659 − Ceratocystis fagacearum CMW 2656 − KM495341 − − Corollospora marina AFTOL-ID 5008 − AY762985 FJ176843 − Corollospora ramulosa PP1232 − AF491276 − − Falcocladium multivesiculatum CBS 120386T EU040217 JF831932 JF831928 − Falcocladium sphaeropedunculatum CBS 111292T JF831938 JF831933 JF831929 − Faurelina elongata CBS 126.78 GU291802 DQ368625 DQ368657 DQ368639 Faurelina fimigena CBS 352.78T MH861149 − − − Graphium fimbriasporum CMW 5605T NR_155108 NG_067529 NG_064882 − Graphium fimbriisporum CMW 5606 AY148180 − AY148172 − Graphium laricis CMW 5601T AY148183 NG_064370 AY148162 − Graphium penicillioides CBS 102632T KY852474 NG_067324 − − Graphium pseudormiticum CMW 503T NR_136962 NG_064371 NG_064881 − Knoxdaviesia capensis AFTOL-ID 1907 − FJ176888 FJ176834 − Knoxdaviesia proteae CBS 486.88T AY372072 AF221011 AY271804 − Knoxdaviesia serotecta CBS 129738T MH865536 MH876970 − − Knoxdaviesia ubusi CBS 129742T MH865540 KM495395 − − Knoxdaviesia wingfieldii CBS 132470T NR_111723 NG_042656 − − Microascus longirostris AFTOL-ID 1237 − − DQ471026 − Nimbospora effusa JKI 5104A − U46892 U46877 DQ836887 Parascedosporium tectonae CBS 127.84T MH861707 EF151332 U43907 − Pseudoscopulariopsis schumacheri CBS 435.86T LM652455 AF400874 − − Synnematotriadelphia stilboidea CBS 101294 MF434783 MF434793 MF434811 MF434802 Synnematotriadelphia stilboidea CBS 221.85T MF434781 MF434792 MF434810 MF434801 Triadelphia disseminata CBS 138592T MF434784 MF434788 MF434806 MF434797 Triadelphia diversa CBS 113.90T MF434782 MF434790 MF434808 MF434799 Triadelphia fusiformis MFLUCC 16-0231T MH777097 MH777098 − − Triadelphia heterospora CBS 222.83T MF434779 MF434789 MF434807 MF434798 Triadelphia hexaformispora TBRC 9288T MK588842 MK588850 MK588840 MK578528 Triadelphia loudetiae CBS 589.77T MF434776 MF434785 MF434803 MF434794 Triadelphia moubasheri AUMC 10746T KY611849 − − − Triadelphia mukdahanensis MFLUCC 23-0049 T OQ871494 OQ871495 OQ883648 OQ871561 Triadelphia pulvinata CBS 590.77T MF434777 MF434786 MF434804 MF434795 Triadelphia pulvinata CBS 744.84 MF434780 MF434787 MF434805 MF434796 Triadelphia romanica CBS 162.79T MF434778 MF434791 MF434809 MF434800 Wardomyces anomalus CBS 299.61 LN850992 LN851044 − − 1 AFTOL-ID: Assembling the Fungal Tree of Life; AUMC: Assiut University Mycological Centre; CBS: Westerdijk Fungal Biodiversity Institute, Utrecht, The Netherlands; CMW: Collection of the Forestry and Agricultural Biotechnology Institute (FABI), University of Pretoria, South Africa; JKI: Culture collection of the Julius Kühn-Institute, Siebeldingen, Germany; MFLUCC: Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; TBRC: Thailand Bioresource Research Center; T: ex-type isolates. 2 ITS: internal transcribed spacer regions 1 and 2 including 5.8S nrRNA gene; LSU: 28S large subunit of the nrRNA gene; SSU: 18S small subunit of the nrRNA gene; RPB2: partial RNA polymerase II second largest subunit gene. Table 1.
Taxa used in this study and their GenBank accession numbers. New sequences are in bold.
-
Conidial forms T. mukdahanensis
(this study)T. heterospora
(Shearer & Crane[23])T. heterospora
(Constantinescu & Samson[24])Type (a):
oblong to cylindrical, brown to dark brown, (1–)2–septate, septum covered with dark band(9.5–)18–25 ×
(3.5–)5–7(–8.5) µm(14.7–)16.2–19(–21.2) ×
3.5–6 µm15–23 × 5–8.5 µm Type (b):
pyriform to broadly clavate, or club-shaped, brown to dark brown, (1–)2–septate, the apical septum covered with broad dark bands, the basal cell subhyaline or light brown to brown20–30 × 7–12 µm – – Type (c):
obclavate to acicular with a narrow long tip, hyaline or yellowish brown, multiseptate– – – Type (d):
fusiform, obclavate with acicular tips, or rounded at the tip, multi-septate, end cells pale brown, central cells brown to dark brown, dark band covering over the central septa– 17.6–23.5 × 8.2–10.6 µm 17.6–23.5 × 8.2–10.6 µm Type (e):
allantoid or reniform, hyaline or pale yellowish, 0–3-septate– – 8.5–15 × 3–5 µm Type (f):
obovate to broadly ellipsoidal, pale brown, unicellular5–9 × 3.5–5 µm – 6–8 × 4–5 µm Table 2.
Conidial forms comparison of Triadelphia mukdahanensis and T. heterospora discussed in this study.
Figures
(2)
Tables
(2)