-
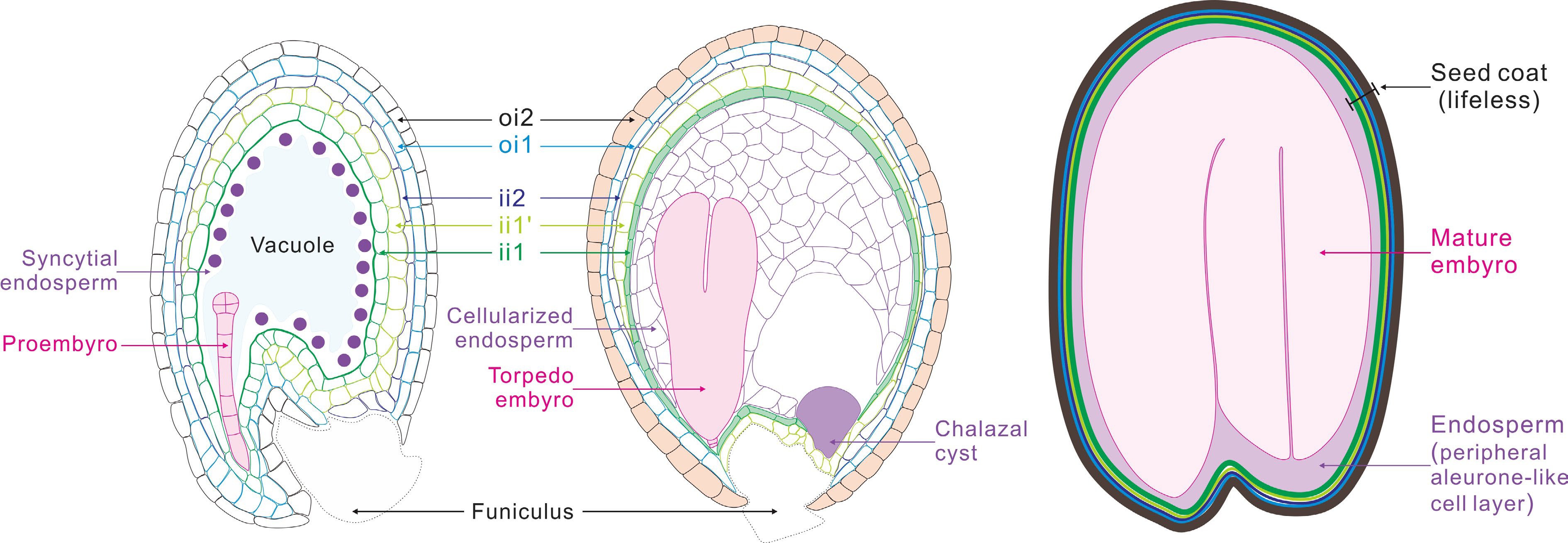
Figure 1.
Schematic representation of seed development in Arabidopsis. After fertilization, the fertilized egg cell (zygote), develops into a proembryo, which then undergoes cell differentiation and organ formation to produce a mature embryo. The primary endosperm cell undertakes continuous mitosis without cell wall formation to form the syncytial endosperm. Then, the endosperm coenocyte initiates cellularization to produce the cellularized endosperm. Finally, the endosperm undergoes programmed cell death (PCD), resulting in only one layer of endosperm cells wrapping the embryo in mature seeds. The two cell layers of outer integument (oi) and three cell layers of inner integument (ii) undergo a period of growth after fertilization, and then adopt distinct cell fates. The innermost endothelial layer (ii1) synthesizes the flavonoid compound proanthocyanidins, and the outermost epidermal layer (oi2) produces and secretes mucilage. By seed maturity, cells of all ii and oi layers are dead.
-
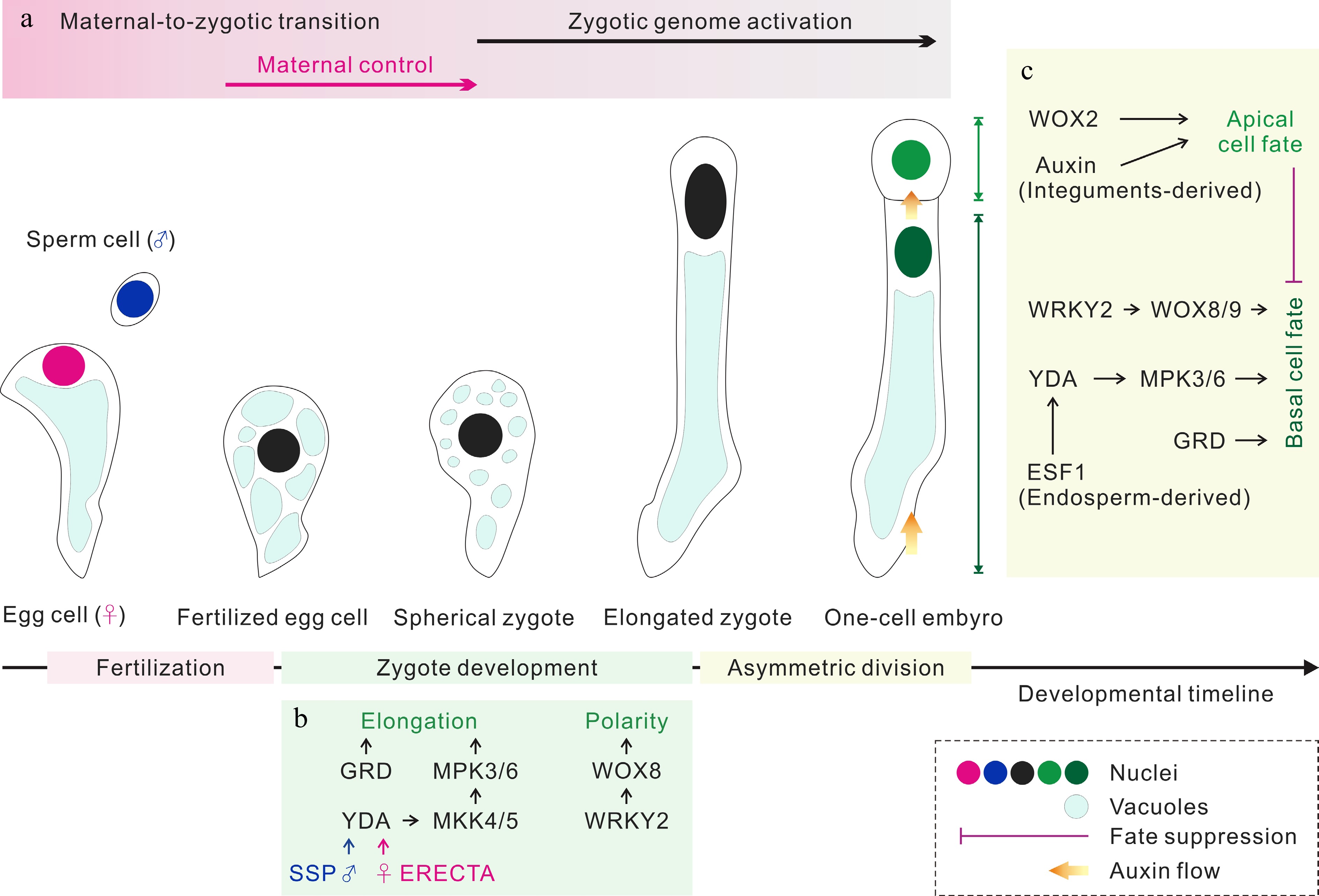
Figure 2.
Zygote development and apical/basal cell fate determination. (a) The parental contributions to the transcriptomes of early embryos. (b) The regulation of zygote development. Both sperm cell-derived SSP and egg cell-provided ERECTA trigger the YDA MAP kinase signaling pathway, which together with RWP-RK Factor GRD to elongate zygote. WRKY2 activates WOX8 to induce the polarity establishment of zygote. (c) The regulation of apical/basal cell fate determination. Following asymmetric zygote division, the auxin response and WOX2 control the apical cell fate, and the WRKY2-WOX8/9 and YDA MAP kinase pathways regulate the basal cell fate. GRD and the endosperm-derived ESF1 regulate basal cell lineage specification through the YDA MAP kinase pathway. In addition, the embryogenic potential of the basal cell lineage is suppressed by the apical cell lineage during normal embryogenesis.
-
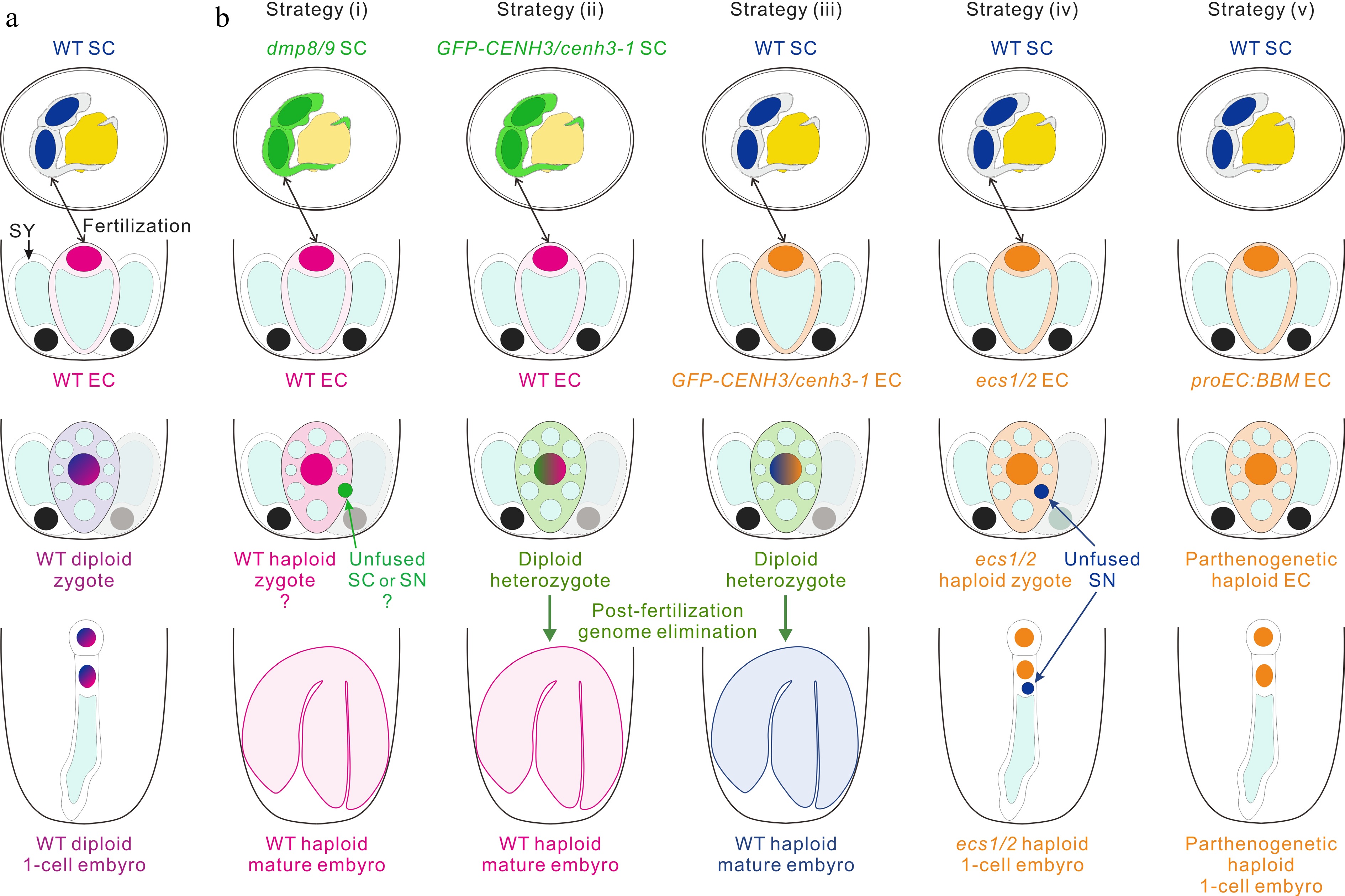
Figure 3.
Strategies for engineering haploid offspring. (a) The haploid sperm cell (SC) and egg cell (EC) fuse to produce a zygote, which then develops into a diploid embryo in wild-type (WT). (b) In vivo haploid embryo development. Strategy (i): the mutants of two sperm-specific genes DMP8 and DMP9 as paternal lines cross with the WT to induce haploid plants. Strategy (ii) and (iii): cenh3-1 plants with the expression of GFP-CENH3 or GFP-tailswap are used to induce haploids by cross of the paternal (ii) and maternal (iii) lines. Strategy (iv): the mutant of two egg cell-specific genes ECS1 and ECS2 as a maternal line and cross with the WT to induce haploid embryogenesis, which may be due to the karyogamy defect. Strategy (v): the egg cell-ectopic expression of Brassica napus BBM trigger parthenogenesis and lead to the formation of haploid plants. SY, synergid cell; SN, sperm cell nucleus.
-

Figure 4.
Some regulatory pathways for the control of central cell and primary endosperm nucleus proliferation. (a), (b) The regulation network inhibiting central cell division before fertilization. PRC2 complex containing FIS1, FIS2, FIE and MSI1 inhibits central cell division and negatively regulate endosperm growth and proliferation. Dysfunction of FIS2 and FIE could activate auxin biosynthesis gene YUC10 and lead to autonomous division of central cell in the absence of fertilization. Similarly, adding the exogenous auxin 2,4-D or co-expressing auxin biosynthesis gene TAA1 and YUC6 in central cell also induce replication of the central cell. In addition, ectopically expression of CYCD3;1 or CYCD7;1 in central cell can induce central cell proliferation. (c) The endosperm proliferation is positively regulated by fertilization. Karyogamy facilitated by CDKA;1 is required for normal endosperm development. The sperm-derived miR159 inhibits its maternal targets, MYB33 and MYB65, in the central cell to promote the initiation of endosperm nuclear divisions. * indicates the ectopic genes expression in the central cell; ** indicates other components of the PRC2 complex.
Figures
(4)
Tables
(0)