-
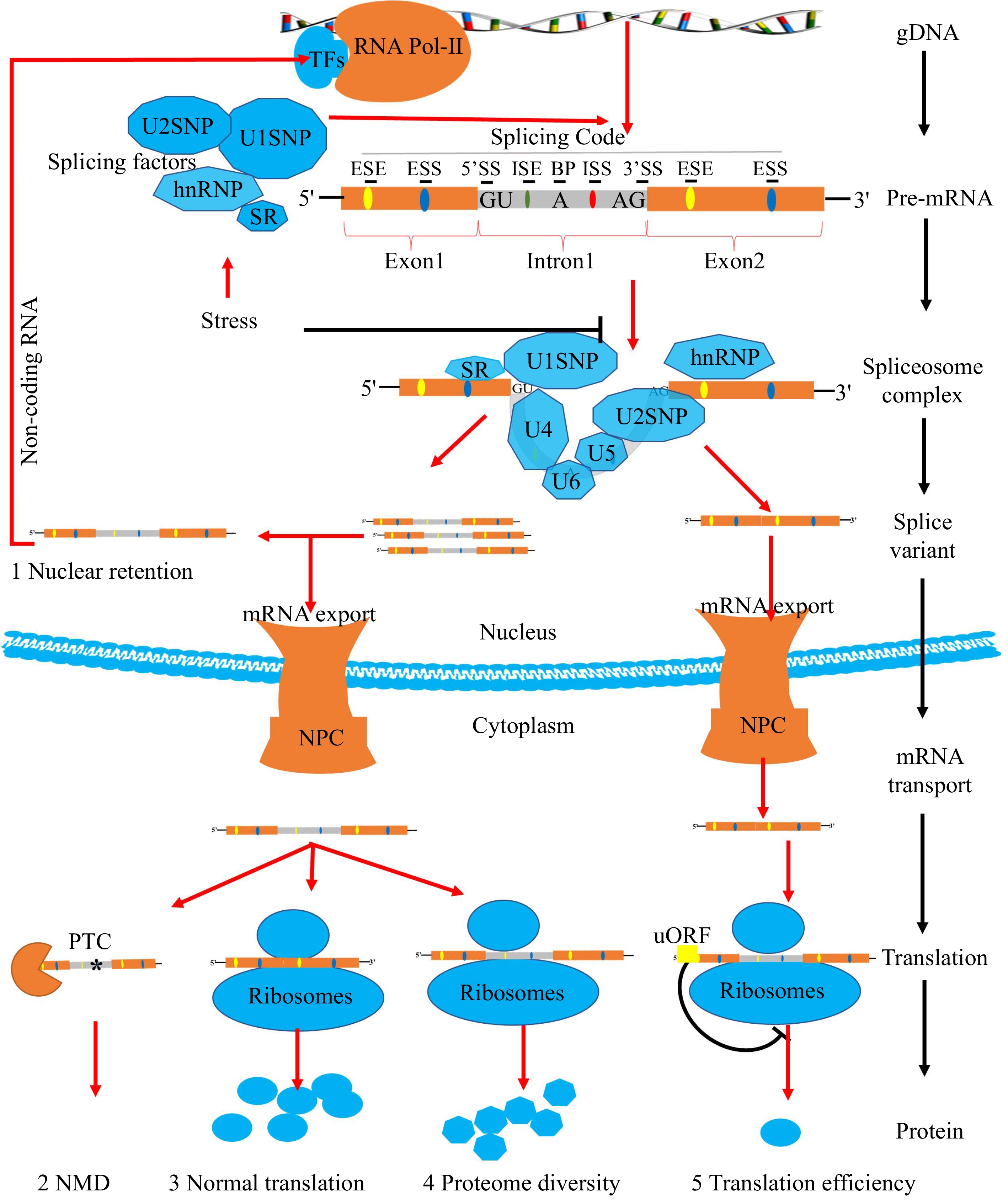
Figure 1.
Pre-mRNA splicing process of plants. In plants, most coding sequences of genes are interrupted by introns. Transcription factors (TFs) recognize promoters and guide the expression of genes. The gDNA is transcribed by RNA polymerase-II (Pol-II) to produce pre-mRNA with both exons and introns. Pre-mRNA owns critical splicing codes, including 5'SS, 3'SS, branch point, polypyrimidine tract, ESEs/ESSs, and ISEs/ISSs. SFs are responsible for recognizing splicing codes. The active spliceosome removes introns, ligating exons of pre-mRNA selectively to produce splicing variants. Stresses have significant effects on AS through SFs. Different splicing variants will have multiple molecular consequences. Firstly, the nuclear pore complex (NPC) will prevent certain intron-containing mRNAs leakage into the cytoplasm and leading to nuclear retention events. Secondly, mRNA transcripts containing premature translation-termination codons (PTCs) with 3'-untranslated regions (3'UTRs) (≥ 350 nt) can be degraded by nonsense-mediated RNA decay (NMD). Finally, the remaining mRNA transcripts will be translated into different spliced proteins by ribosome with different translation efficiency due to the presence of (upstream open reading frames) uORFs.
-
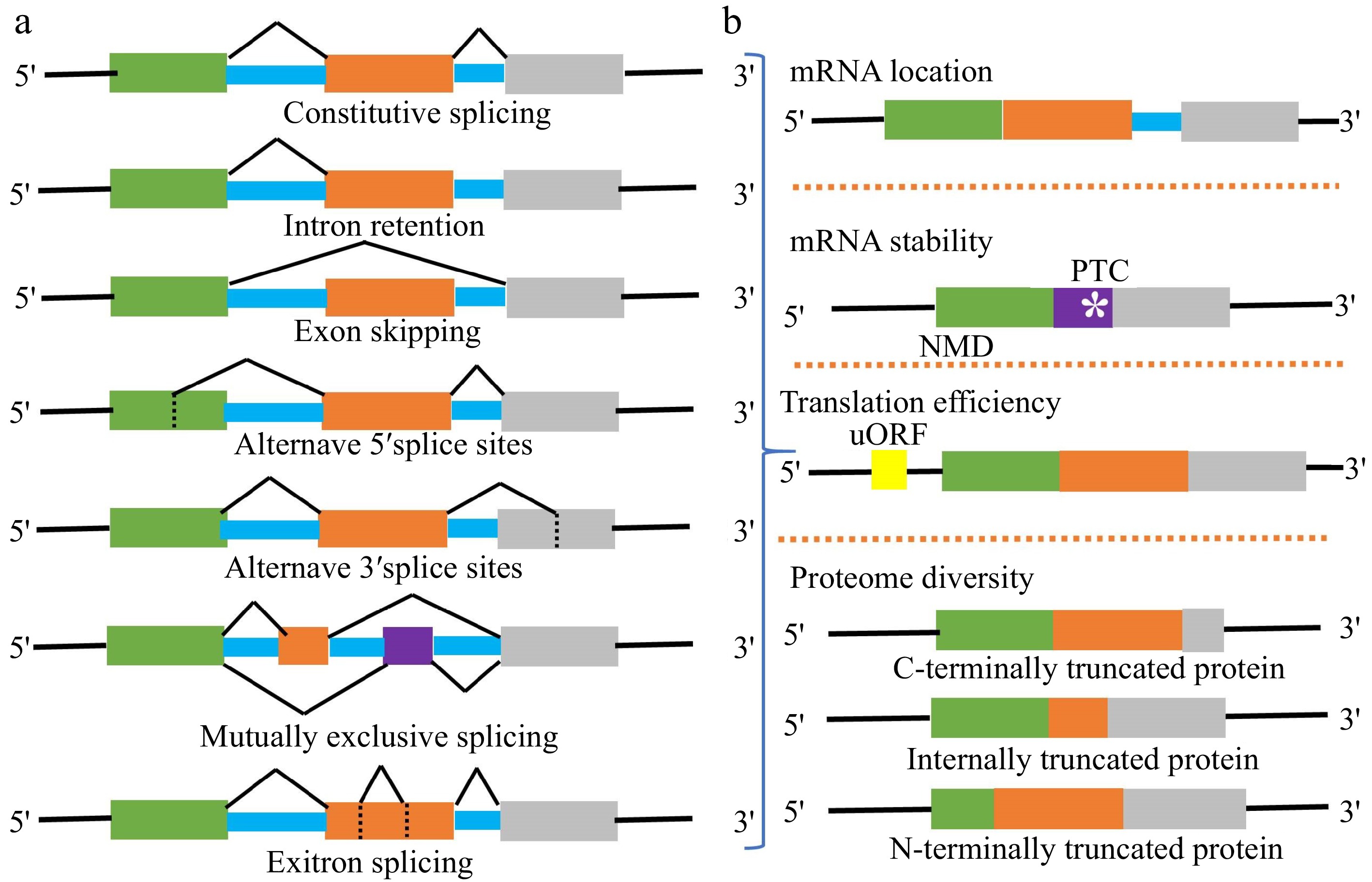
Figure 2.
(a) Major types of AS events in plants include constitutive splicing (CS), intron retention (IR), exon skipping (ES), alternative 5' splice sites (A5SS), alternative 3' splice sites (A3SS), mutually exclusive splicing (MES), and exitron splicing (EIS). Usually, an mRNA splicing variant is formed by one or more AS events. (b) Different AS event combinations lead to generate multiple splicing variants from one gene locus. The subcellular location, stability, translation efficiency, and coding sequence of splicing variants are different.
-
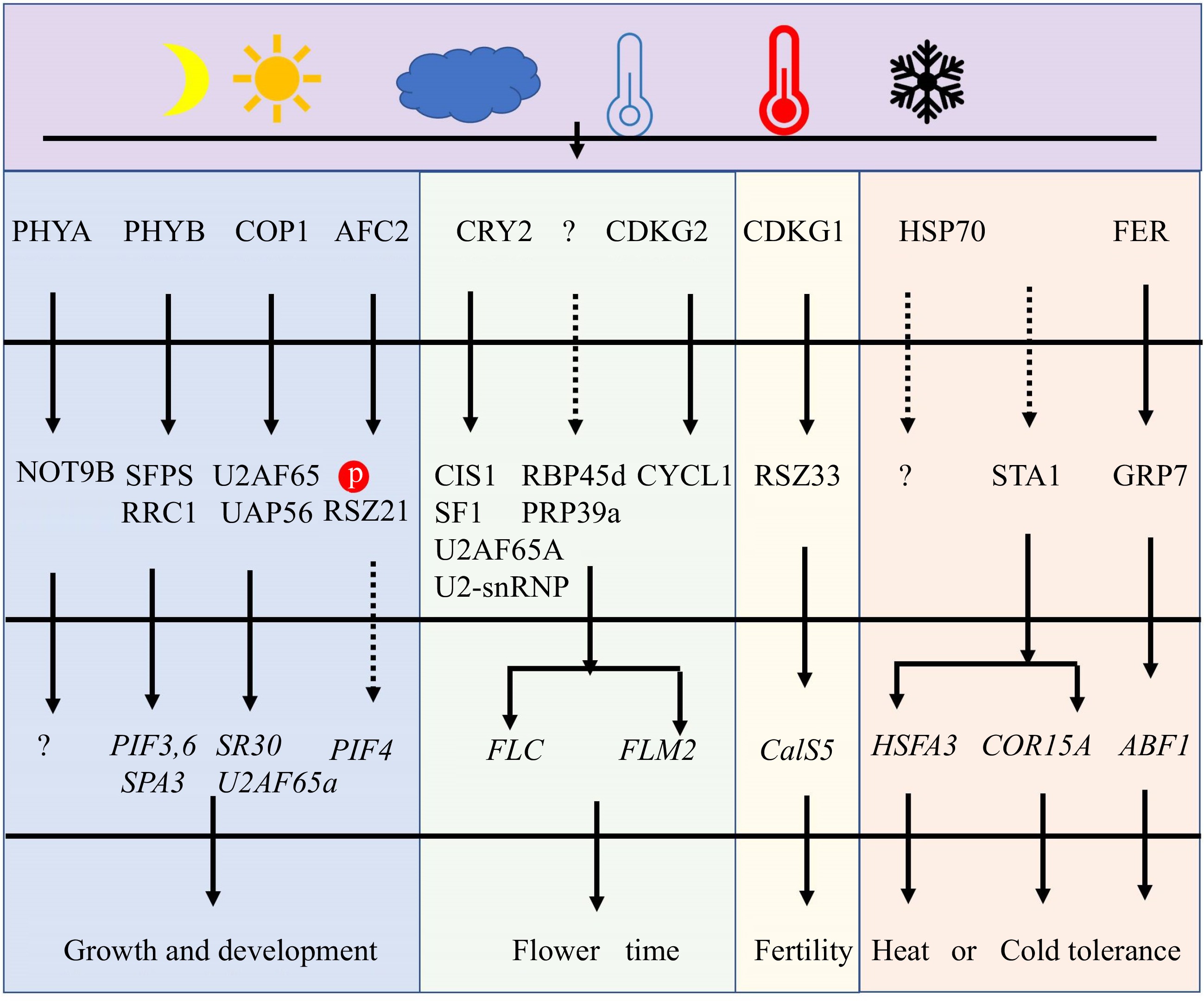
Figure 3.
The major pathway for regulating pre-mRNA AS under light and temperature stresses. At first, some SFs could act as sensors to perceive the changes in light and temperatures. Secondly, those sensors will phosphorylate or interact with other SFs and change their location and activity. These modifications of SFs will affect AS by controlling spliceosome function. Finally, plant growth, development, flower, fertility, and tolerance are regulated under adverse light and temperature conditions.
-
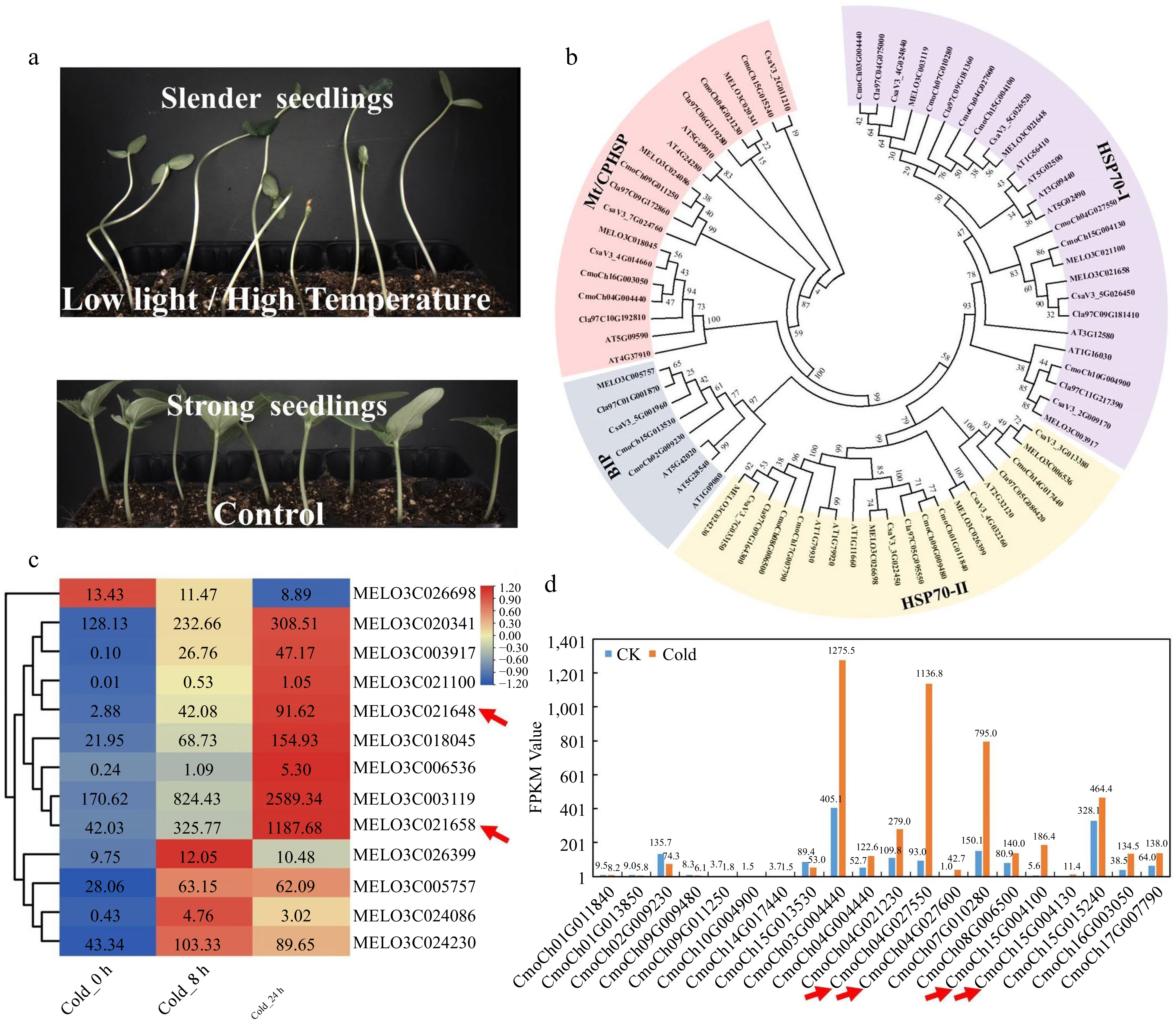
Figure 4.
Expression patterns analysis of the HSP70 gene family members in cucurbit crops. (a) Low light or high temperature stress causes slender cucumber seedlings in the breeding greenhouse. (b) The phylogenetic tree of the HSP70 gene family from cucumber, pumpkin, watermelon, tomato, melon, and Arabidopsis. (c) Heatmap of melon HSP70 gene family after 8 h and 24 h cold treatment. (d) Gene expression patterns of the pumpkin HSP70 gene family after 24 h cold treatment.
-
Annotation Name Species Gene ID Function Reference DEAD-box RNA heatlicases RH42 Rice Os08G0159900 Cold Tolerance Lu et al.[65] U1 snRNP Prp39a Arabidopsis AT1G04080 Temperature-induced flowering time Chang et al.[73] hnRNPs RBP45d Arabidopsis AT5G19350 Temperature-induced flowering time Chang et al.[73] Phytochrome CRY2 Arabidopsis AT1G04400 Blue-light regulation of thermosensory flowering Zhao et al.[74] mRNA binding SF1/BBP Arabidopsis AT5G51300 Temperature-induced flowering time Lee et al.[71] hnRNP A/B family UBA2c Arabidopsis AT3G15010 Light and temperature-induced flowering time Zhao et al.[72] Core proteins (Sm) SmE-b/PCP/SME1 Arabidopsis AT2G18740 Arrest of growth and male sterile at
low temperatureCapovilla et al.[17] U1 snRNP Luc7a Arabidopsis AT3G03340 Low temperature resulted in arrest
of growthAmorim et al.[63] U1 snRNP Luc7b Arabidopsis AT5G17440 Low temperature resulted in arrest
of growthAmorim et al.[63] U1 snRNP Luc7-rl Arabidopsis AT5G51410 Low temperature resulted in arrest
of growthAmorim et al.[63] Glycine rich protein GRPs-7 Arabidopsis AT2G30560 Cold tolerance Kim et al.[62] DEAD box RNA helicase Prp5-1b/RCF1 Arabidopsis AT1G20920 Cold tolerance and cold acclimion Guan et al.[59] U5 snRNP U5-200-2b/Brr2b Arabidopsis AT2G42270 Cold tolerance and cold acclimion Guan et al.[59] DEAD-box RNA helicase U2B"a/U2B"-LIKE Arabidopsis AT1G06960 Cold tolerance and cold acclimion Calixto et al.[64] SR protein kinase AFC3/AME3 Arabidopsis AT4G32660 Cold tolerance and cold acclimion L. Savitch, unpublisheatd data 17S U2 snRNP GEMIN2 Arabidopsis AT1G54380 Cold tolerance and cold acclimion Schlaen et al.[61] U5 snRNP U5-102KD/STA1 Arabidopsis AT4G03430 Cold tolerance and cold acclimion;
heat tolerance and heat acclimionKim et al.[66];
Guan et al.[59]Inositol-requiring enzyme-1 IRE1b Arabidopsis AT5G24360 Heat stress responses Deng et al.[39]; Moreno et al.[38] Inositol-requiring enzyme-1 IRE1a Arabidopsis AT2G17520 Heat stress responses Deng et al.[39]; Moreno et al.[38] hnRNP HOS5/RCF3 Arabidopsis AT5G53060 Heat stress tolerance Guan et al.[60] HSP70-3 HSP70-3 Arabidopsis AT3G09440 Heat stress tolerance Song et al.[56] HSP70-4 HSP70-4 Arabidopsis AT3G12580 Heat stress tolerance Wang et al.[57] HSC70-1 HSC70-1 Arabidopsis AT5G02500 Heat stress tolerance Tiwari et al.[58] Cyclin-dependent kinase CDCL2p110-1 Arabidopsis AT1G67580 High temperature-induced male fertility defects Cavallari et al.[70]; Zheng et al.[69]; Nibau et al.[68] Cyclin-dependent kinase CDCL2p110-2/CDKG1 Arabidopsis AT5G63370 High temperature induce fertility defects Cavallari et al.[70]; Zheng et al.[69]; Nibau et al.[68] SR protein Kinase AFC2 Arabidopsis AT4G24740 Thermosensors and thermomorphogenesis Lin et al.[88] Phytochrome PHYB Arabidopsis AT2G18790 Thermosensors and thermomorphogenesis;
photoreceptors and photomorphogenesisJung et al.[48]; Shikata et al.[12,51]; Hartmann et al.[52]; Xin et al.[35,50] Phytochrome PHYA Arabidopsis AT1G09570 Photomorphogenesis Shikata et al.[12]; Schwenk et al.[53] E3 ubiquitin ligase COP1 Arabidopsis AT2G32950 Photomorphogenesis Li et al.[54] 17S U2 associated proteins SR140-1/RRC1 Arabidopsis AT5G25060 Photomorphogenesis Shikata et al.[12,51]; Hartmann et al.[52]; Xin et al.[35,50] Table 1.
Splicing factors involved in light and temperature stress adaption.
Figures
(4)
Tables
(1)