-
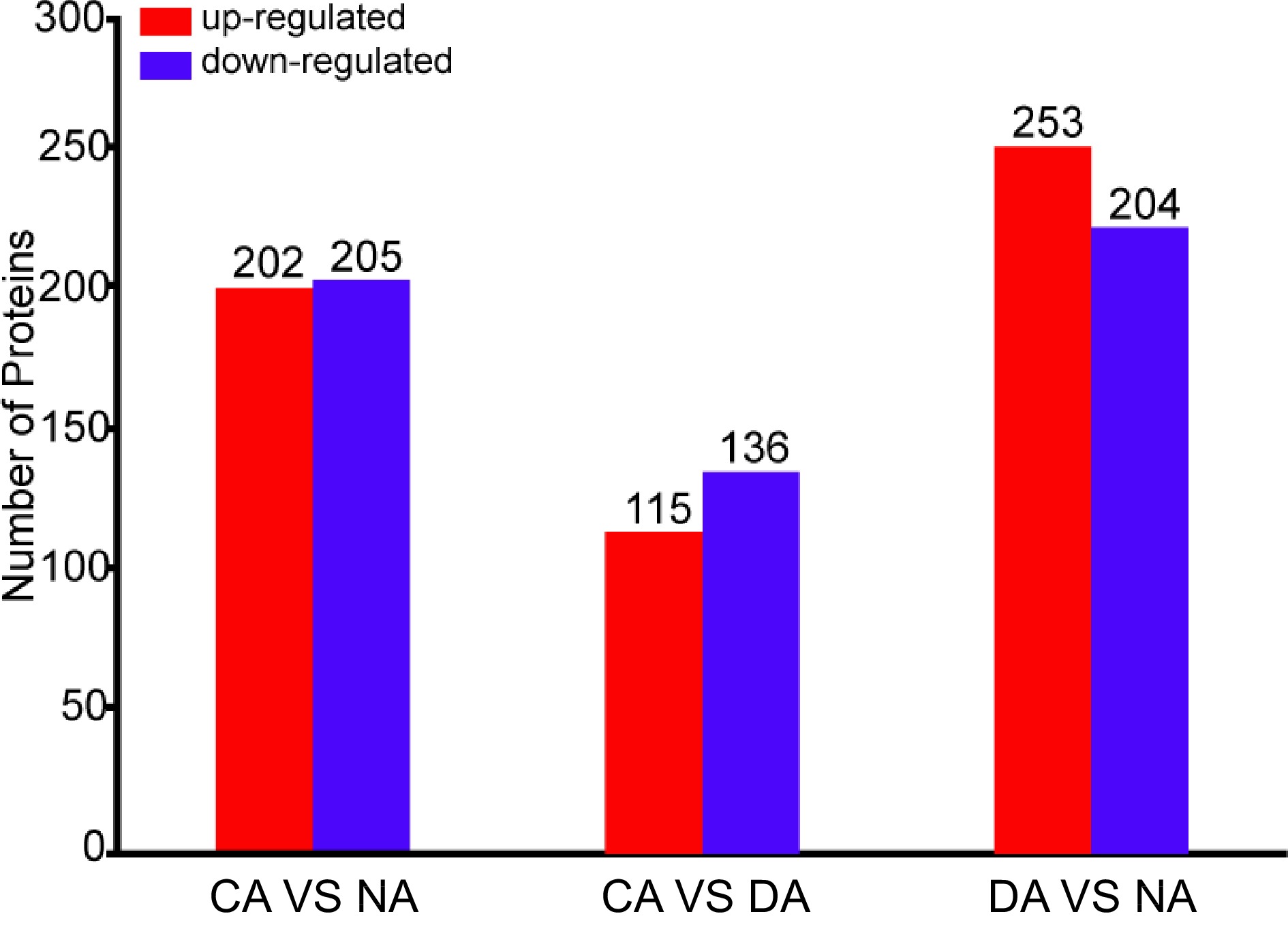
Figure 1.
Number of differentially accumulated proteins among different samples.
-
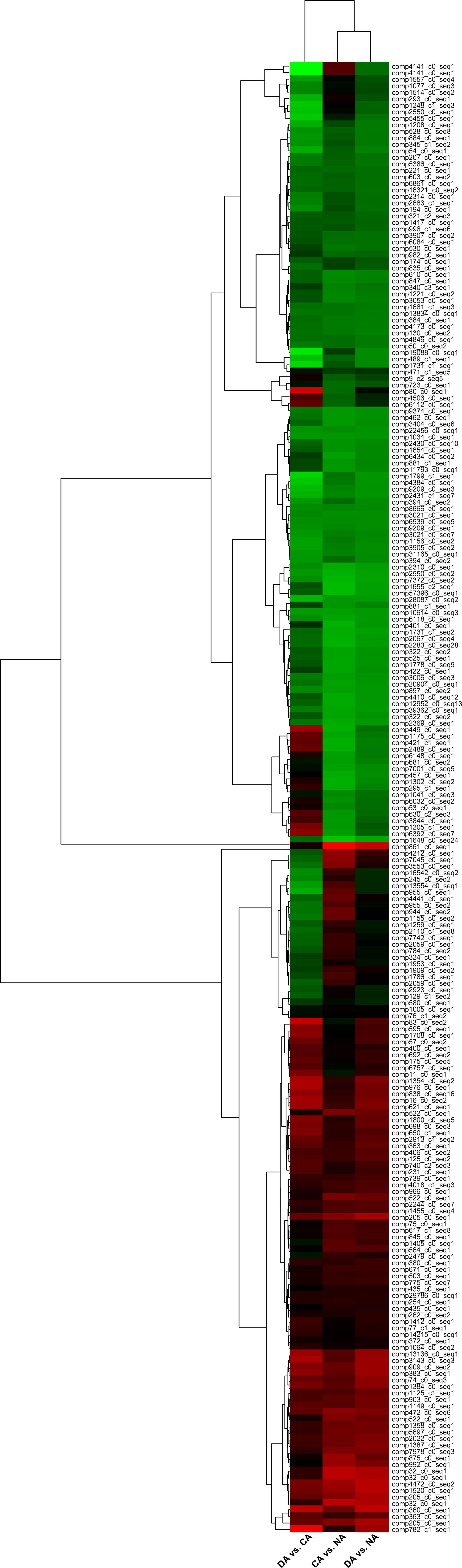
Figure 2.
Hierachical clustering of proteins showing different abundance profiles across different samples. The data were transferred using log2.
-
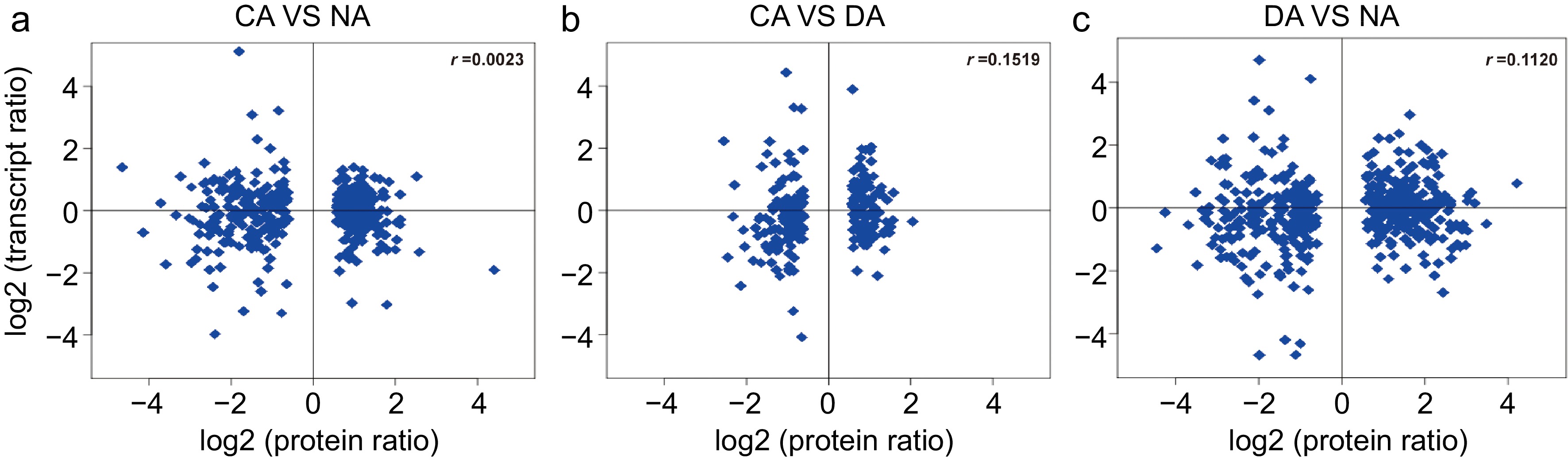
Figure 3.
Correlation analysis of transcript (log2 FPKM value) and protein (log2 iTRAQ value) among different samples. (a) CA vs NA; (b) CA vs DA; (c) DA vs NA.
-
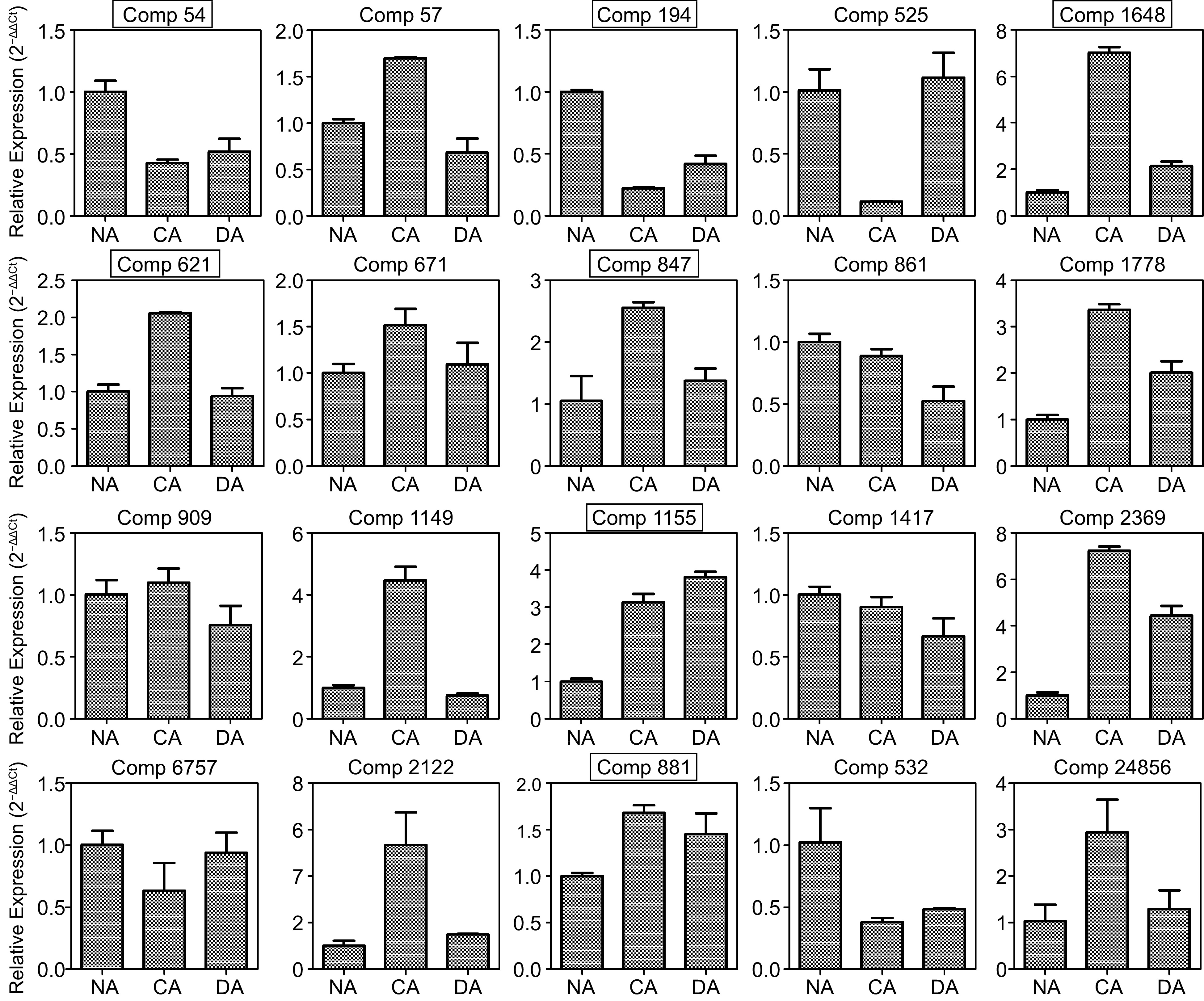
Figure 4.
Analysis of transcript levels of the selected proteins among NA, CA and DA stages by qRT-PCR. Those genes which showed similar patterns are marked using black boxes. All data are the mean ± SD (n = 3).
-
GO term NA vs CA CA vs DA DA vs NA Cellular component Extracellular region Cell wall Extracellular region Plastid stroma Plastid part Cell wall Cytoplasmic vesicle Plastid stroma Plastid envelope Vesicle Thylakoid light-harvesting complex External encapsulating structure Chloroplast thylakoid membrane Organelle envelope Light-harvesting complex Envelope Plastid thylakoid membrane Microbody Chromosome Membrane part Molecular function Ammonia ligase activity Adenylyltransferase activity Binding Acid-ammonia (or amide) ligase activity Racemase and epimerase activity Protein kinase activity Oxidoreductase activity, acting on the
CH-NH2 group of donors, disulfide as
acceptorO-acyltransferase activity Ammonia ligase activity Ligase activity, forming carbon-nitrogen
bondsRacemase and epimerase activity, acting
on carbohydrates and derivativesAcid-ammonia (or amide) ligase activity Binding Biological process Ribosome biogenesis Electron transport chain Electron transport chain Glutamine metabolic process Oxidation reduction Oxidation reduction Cellular carbohydrate metabolic process Nucleoside phosphate metabolic process Photosynthetic electron transport chain Reproductive developmental process Water-soluble vitamin metabolic process Nucleotide metabolic process Reproductive process Nucleobase, nucleoside and nucleotide metabolic process Generation of precursor metabolites and energy Glycine metabolic process Photosynthetic electron transport chain Ribosome biogenesis Cellular amino acid metabolic process Nucleotide metabolic process Nucleoside phosphate metabolic process Cellular amine metabolic process Seed germination Glutamine metabolic process Sulfur amino acid metabolic process Mucilage metabolic process Nucleobase, nucleoside and nucleotide metabolic process Reproductive structure development Glucan metabolic process Negative regulation of molecular function Carbohydrate metabolic process Vitamin metabolic process Protein complex assembly Reproduction NADP metabolic process Oxidoreduction coenzyme metabolic process Amine metabolic process NADPH regeneration Tissue development Respiratory electron transport chain Nicotinamide metabolic process Pyridine nucleotide metabolic process Flower development Alkaloid metabolic process Cellular macromolecular complex assembly Organic acid catabolic process Nucleobase, nucleoside, nucleotide and nucleic acid metabolic process Cellular protein complex assembly Carboxylic acid catabolic process Cellular glucan metabolic process Nicotinamide nucleotide metabolic process Response to chemical stimulus Photosynthesis, light reaction Protein complex biogenesis Response to inorganic substance Purine nucleotide metabolic process Cellular component biogenesis Energy derivation by oxidation of organic compounds Oxidoreduction coenzyme metabolic
processCellular component organization Organic acid metabolic process Pyridine nucleotide metabolic process Sulfur metabolic process Carboxylic acid metabolic process Nicotinamide nucleotide metabolic process Primary metabolic process Cellular ketone metabolic process Cellular polysaccharide metabolic process Photosynthesis Oxoacid metabolic process Glycogen metabolic process Purine nucleotide metabolic process Sulfur amino acid biosynthetic process Energy reserve metabolic process Fatty acid metabolic process Serine family amino acid metabolic process Photosynthesis Photosynthesis, light reaction Generation of precursor metabolites and energy Plastid membrane organization Stomatal movement Membrane organization Table 1.
Gene Ontology (GO) enrichment analysis of differentially accumulated protein species among the comparisons between sample NA, CA and DA ( p -value < 0.05).
-
Number Pathway Count Pathway ID Total
(1,015)CA vs NA
(319)CA vs DA
(196)DA vs NA
(362)1 Metabolic pathways 458 149 91 168 ko01100 2 Biosynthesis of secondary metabolites 247 75 33 74 ko01110 3 Microbial metabolism in diverse environments 190 70 32 73 ko01120 4 Ribosome 83 24 17 27 ko03010 5 Carbon fixation in photosynthetic organisms 71 33 13 31 ko00710 6 Glycolysis / Gluconeogenesis 65 22 5 25 ko00010 7 Methane metabolism 53 17 8 21 ko00680 8 Starch and sucrose metabolism 52 19 10 24 ko00500 9 Protein processing in endoplasmic reticulum 49 17 11 15 ko04141 10 Lysosome 49 11 10 16 ko04142 11 Pyruvate metabolism 48 18 7 20 ko00620 12 Amino sugar and nucleotide sugar metabolism 43 11 8 14 ko00520 13 Photosynthesis 43 13 13 17 ko00195 14 Glutathione metabolism 36 7 7 14 ko00480 15 Phenylpropanoid biosynthesis 34 11 7 13 ko00940 16 Alanine, aspartate and glutamate metabolism 31 12 7 11 ko00250 17 Citrate cycle (TCA cycle) 31 10 3 10 ko00020 18 Plant-pathogen interaction 30 12 7 15 ko04626 19 Pentose phosphate pathway 30 13 6 12 ko00030 20 Glycine, serine and threonine metabolism 29 13 6 13 ko00260 21 Antigen processing and presentation 29 10 5 8 ko04612 22 Oxidative phosphorylation 28 11 8 12 ko00190 23 Glyoxylate and dicarboxylate metabolism 26 12 5 11 ko00630 24 Purine metabolism 25 7 4 7 ko00230 25 Huntington's disease 25 9 4 5 ko05016 26 Phenylalanine metabolism 25 10 8 10 ko00360 27 Fructose and mannose metabolism 25 8 8 7 ko00051 28 Peroxisome 23 7 4 9 ko04146 29 Galactose metabolism 22 4 5 8 ko00052 30 Arginine and proline metabolism 22 7 3 5 ko00330 31 Phagosome 21 3 5 11 ko04145 32 Nitrogen metabolism 21 13 10 13 ko00910 33 Cysteine and methionine metabolism 19 7 1 6 ko00270 34 Spliceosome 19 6 3 6 ko03040 35 Other glycan degradation 19 6 3 7 ko00511 36 Alzheimer's disease 18 8 3 8 ko05010 37 Valine, leucine and isoleucine degradation 17 4 1 7 ko00280 38 Parkinson's disease 16 5 3 3 ko05012 39 Butanoate metabolism 16 5 2 5 ko00650 40 Ascorbate and aldarate metabolism 16 7 3 4 ko00053 41 RNA degradation 16 5 2 6 ko03018 42 Pentose and glucuronate interconversions 15 7 4 7 ko00040 43 Glycerolipid metabolism 15 6 3 7 ko00561 44 Endocytosis 13 5 4 3 ko04144 45 Toxoplasmosis 13 5 3 3 ko05145 46 Propanoate metabolism 13 5 2 3 ko00640 47 Phenylalanine, tyrosine and tryptophan biosynthesis 13 3 3 2 ko00400 48 Aminoacyl-tRNA biosynthesis 13 4 1 4 ko00970 49 Cyanoamino acid metabolism 13 8 2 6 ko00460 50 Renin-angiotensin system 13 3 5 6 ko04614 51 Aminobenzoate degradation 12 4 4 6 ko00627 52 Inositol phosphate metabolism 12 4 2 4 ko00562 53 RNA transport 12 7 2 7 ko03013 54 Valine, leucine and isoleucine biosynthesis 12 3 1 2 ko00290 55 Selenoamino acid metabolism 11 3 3 4 ko00450 56 Tyrosine metabolism 11 6 3 4 ko00350 57 Tropane, piperidine and pyridine alkaloid biosynthesis 11 3 5 1 ko00960 58 Proteasome 11 2 2 ko03050 59 Reductive carboxylate cycle (CO2 fixation) 11 1 3 ko00720 60 Tryptophan metabolism 10 3 1 5 ko00380 61 Two-component system 10 6 5 8 ko02020 62 MAPK signaling pathway 10 4 3 2 ko04010 63 Fatty acid metabolism 10 4 6 ko00071 64 Porphyrin and chlorophyll metabolism 9 2 2 ko00860 65 beta-Alanine metabolism 9 3 1 2 ko00410 66 Glycosphingolipid biosynthesis - globo series 9 5 1 5 ko00603 67 alpha-Linolenic acid metabolism 9 1 1 4 ko00592 68 Terpenoid backbone biosynthesis 9 1 1 2 ko00900 69 Prion diseases 9 7 2 3 ko05020 70 Type I diabetes mellitus 9 3 2 ko04940 71 Pyrimidine metabolism 9 1 1 ko00240 72 One carbon pool by folate 9 5 3 4 ko00670 73 Flavonoid biosynthesis 8 4 1 2 ko00941 74 Chagas disease 8 3 4 ko05142 75 Lysine biosynthesis 8 1 1 2 ko00300 76 Insulin signaling pathway 8 2 1 2 ko04910 77 Chloroalkane and chloroalkene degradation 8 2 1 2 ko00625 78 Carotenoid biosynthesis 8 2 1 3 ko00906 79 Pathogenic Escherichia coli infection 8 1 4 6 ko05130 80 Proximal tubule bicarbonate reclamation 8 1 2 2 ko04964 81 Glycosphingolipid biosynthesis - ganglio series 7 2 2 3 ko00604 82 PPAR signaling pathway 7 2 3 ko03320 83 Limonene and pinene degradation 7 3 3 ko00903 84 Glycosaminoglycan degradation 7 2 2 3 ko00531 85 Metabolism of xenobiotics by cytochrome P450 7 3 4 3 ko00980 86 Sphingolipid metabolism 7 3 3 4 ko00600 87 Glycerophospholipid metabolism 7 3 1 3 ko00564 88 Vibrio cholerae infection 7 2 4 ko05110 89 Drug metabolism - cytochrome P450 7 3 4 3 ko00982 90 Lysine degradation 7 3 3 ko00310 91 Amyotrophic lateral sclerosis (ALS) 7 4 2 4 ko05014 92 Calcium signaling pathway 6 1 1 ko04020 93 NOD-like receptor signaling pathway 6 3 2 3 ko04621 94 Protein digestion and absorption 6 4 2 ko04974 95 Isoquinoline alkaloid biosynthesis 6 3 2 1 ko00950 96 Pathways in cancer 6 3 2 3 ko05200 97 Prostate cancer 6 3 2 3 ko05215 98 Neurotrophin signaling pathway 6 2 4 ko04722 99 Folate biosynthesis 6 1 3 3 ko00790 100 Ubiquinone and other terpenoid-quinone biosynthesis 6 2 3 ko00130 101 Protein export 5 2 2 ko03060 102 Photosynthesis - antenna proteins 5 2 2 2 ko00196 103 Histidine metabolism 5 2 2 ko00340 104 Riboflavin metabolism 5 4 2 2 ko00740 105 Sulfur metabolism 5 1 2 1 ko00920 106 Streptomycin biosynthesis 5 2 ko00521 107 Benzoate degradation 5 2 3 ko00362 108 Bisphenol degradation 5 1 1 1 ko00363 109 MAPK signaling pathway - yeast 5 1 2 2 ko04011 110 Arachidonic acid metabolism 5 1 1 1 ko00590 111 Progesterone-mediated oocyte maturation 5 3 1 2 ko04914 112 Type II diabetes mellitus 5 1 1 ko04930 113 Biosynthesis of ansamycins 5 4 2 4 ko01051 114 Chlorocyclohexane and chlorobenzene degradation 5 2 2 ko00361 115 Novobiocin biosynthesis 5 2 1 1 ko00401 116 Fatty acid biosynthesis 4 1 2 ko00061 117 Polycyclic aromatic hydrocarbon degradation 4 3 1 1 ko00624 118 Linoleic acid metabolism 4 1 ko00591 119 Systemic lupus erythematosus 4 1 2 ko05322 120 Bacterial invasion of epithelial cells 4 2 1 ko05100 121 Regulation of actin cytoskeleton 4 2 2 3 ko04810 122 Biosynthesis of unsaturated fatty acids 4 2 ko01040 123 Geraniol degradation 4 1 2 ko00281 124 Focal adhesion 4 1 2 1 ko04510 125 Collecting duct acid secretion 4 3 ko04966 126 Pantothenate and CoA biosynthesis 4 1 ko00770 127 Epithelial cell signaling in Helicobacter pylori infection 4 3 ko05120 128 Oocyte meiosis 4 1 3 ko04114 129 Cell cycle 4 1 2 ko04110 130 Fluorobenzoate degradation 3 1 ko00364 131 Gap junction 3 2 3 ko04540 132 Toll-like receptor signaling pathway 3 1 1 ko04620 133 N-Glycan biosynthesis 3 1 ko00510 134 Taurine and hypotaurine metabolism 3 1 1 1 ko00430 135 Amoebiasis 3 1 ko05146 136 mRNA surveillance pathway 3 2 2 ko03015 137 Caprolactam degradation 3 1 1 1 ko00930 138 Carbohydrate digestion and absorption 3 2 2 ko04973 139 Ether lipid metabolism 3 1 1 1 ko00565 140 Toluene degradation 3 1 ko00623 141 Shigellosis 3 1 2 2 ko05131 142 Tight junction 3 2 1 ko04530 143 Phototransduction - fly 3 2 2 ko04745 144 Vitamin B6 metabolism 3 1 1 ko00750 145 Drug metabolism - other enzymes 2 ko00983 146 Leukocyte transendothelial migration 2 2 1 ko04670 147 Leishmaniasis 2 1 1 ko05140 148 D-Glutamine and D-glutamate metabolism 2 ko00471 149 Phosphatidylinositol signaling system 2 1 ko04070 150 Cell cycle - Caulobacter 2 1 1 1 ko04112 151 Naphthalene degradation 2 1 1 ko00626 152 Bacterial secretion system 2 1 ko03070 153 Ethylbenzene degradation 2 1 ko00642 154 C5-Branched dibasic acid metabolism 2 ko00660 155 Base excision repair 2 1 1 ko03410 156 Fc gamma R-mediated phagocytosis 2 2 1 2 ko04666 157 Viral myocarditis 2 2 1 ko05416 158 GnRH signaling pathway 2 1 1 2 ko04912 159 Flavone and flavonol biosynthesis 2 2 2 ko00944 160 Arrhythmogenic right ventricular cardiomyopathy (ARVC) 2 2 1 ko05412 161 Dilated cardiomyopathy 2 2 1 ko05414 162 Apoptosis 2 1 1 ko04210 163 ECM-receptor interaction 2 1 ko04512 164 Hypertrophic cardiomyopathy (HCM) 2 2 1 ko05410 165 DDT degradation 2 2 1 ko00351 166 Ubiquitin mediated proteolysis 2 1 1 ko04120 167 Adherens junction 2 2 1 ko04520 168 Melanogenesis 1 1 ko04916 169 Vascular smooth muscle contraction 1 1 ko04270 170 Basal transcription factors 1 1 1 ko03022 171 Lipopolysaccharide biosynthesis 1 ko00540 172 Nucleotide excision repair 1 ko03420 173 Stilbenoid, diarylheptanoid and gingerol biosynthesis 1 1 1 ko00945 174 Steroid biosynthesis 1 ko00100 175 RIG-I-like receptor signaling pathway 1 ko04622 176 Cardiac muscle contraction 1 1 1 ko04260 177 Phototransduction 1 1 ko04744 178 Sulfur relay system 1 1 1 ko04122 179 Gastric acid secretion 1 1 ko04971 180 Retinol metabolism 1 1 1 ko00830 181 Circadian rhythm - plant 1 ko04712 182 Mismatch repair 1 ko03430 183 Salivary secretion 1 1 ko04970 184 Benzoxazinoid biosynthesis 1 ko00402 185 Fatty acid elongation in mitochondria 1 ko00062 186 Notch signaling pathway 1 1 1 1 ko04330 187 Thiamine metabolism 1 1 1 ko00730 188 DNA replication 1 ko03030 189 Indole alkaloid biosynthesis 1 ko00901 190 Synthesis and degradation of ketone bodies 1 1 1 ko00072 191 Long-term potentiation 1 1 ko04720 192 Diterpenoid biosynthesis 1 ko00904 193 SNARE interactions in vesicular transport 1 ko04130 194 Glioma 1 1 ko05214 195 Olfactory transduction 1 1 ko04740 Table 2.
Pathway analysis of total proteins and enriched proteins in the comparisons among different samples.
Figures
(4)
Tables
(2)