-
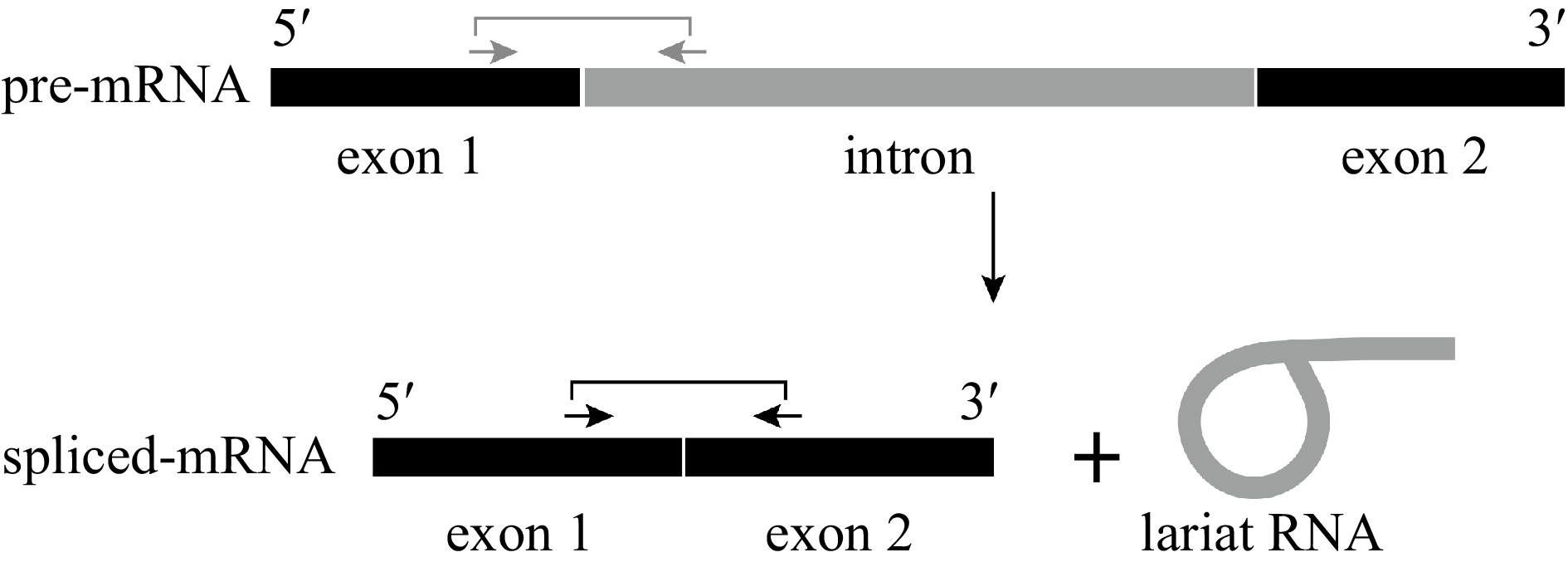
Figure 1.
Process of mRNA splicing. RNA splicing involves cutting an intron (gray) from pre-mRNA and joining together the two neighboring exons (black). The spliced exons form the functional RNA, and the intron is usually degraded. Arrows indicate the positions of the primers used to detect the pre-mRNA (gray) and spliced mRNA (black).
-
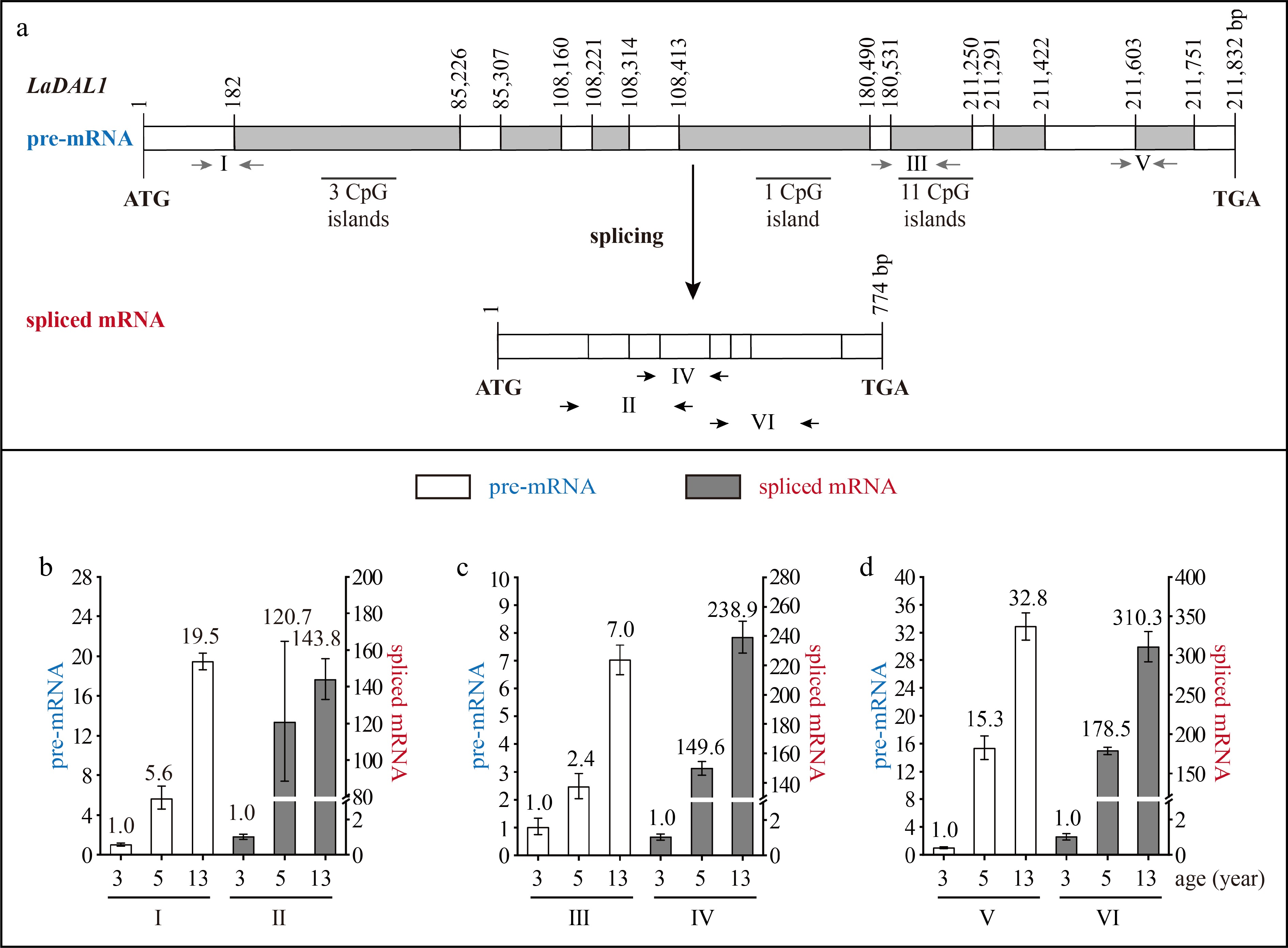
Figure 2.
Structure and expression pattern of LaDAL1. (a) Schematic representation of the gene structure of LaDAL1. White indicates exon; gray indicates intron; gray arrows indicate the positions of the primers used to measure the pre-mRNA; and black and horizontal arrows indicate the positions of the primers used to measure the spliced mRNA. After the prediction of DNA methylation, the CpG islands were revealed. (b)–(d) Expression patterns of LaDAL1 pre-mRNA and spliced mRNA during tree aging detected by three different primer pairs. The lateral branches of 3-, 5-, and 13-year-old active Larix kaempferi trees (n ≥ 6, sampled on 4 July 2019) were used to examine the expression patterns, which were assayed by qRT-PCR with LaEF1A1 as the internal control. The capitalized Roman numerals (I–VI) in panels (a)–(d) represent the different primers. The p-values of the differences between 5- and 3-year-old trees were calculated. One-way ANOVA Duncan’s test was used for statistical analysis.
-
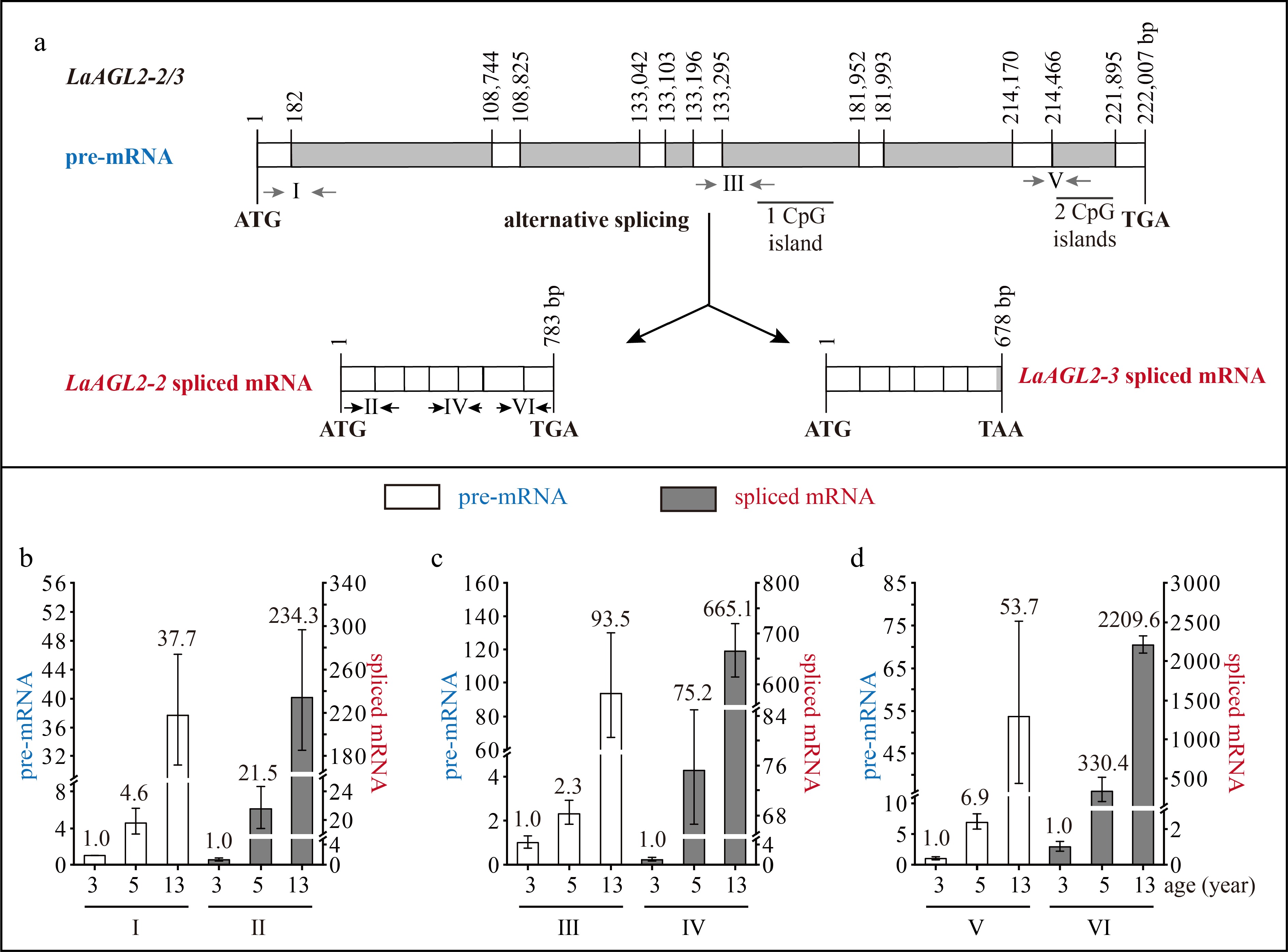
Figure 3.
Structure and expression pattern of LaAGL2-2/3. (a) Schematic representation of the gene structure of LaAGL2-2/3. White indicates exon; gray indicates intron; gray arrows indicate the positions of the primers used to measure the pre-mRNA; and black and horizontal arrows indicate the positions of the primers used to measure the spliced mRNA. After the prediction of DNA methylation, the CpG islands were revealed. (b)–(d) Expression patterns of LaAGL2-2/3 (b, c) and LaAGL2-2 (d) pre-mRNA and spliced mRNA during tree aging detected by three different primer pairs. The lateral branches of 3-, 5-, and 13-year-old active Larix kaempferi trees (n ≥ 6, sampled on 4 July 2019) were used to detect the expression patterns, which were assayed by qRT-PCR with LaEF1A1 as the internal control. The capitalized Roman numerals (I–VI) in panels (a)–(d) represent the different primers. The p-values of the differences between 5- and 3-year-old trees were calculated. One-way ANOVA Duncan’s test was used for statistical analysis.
-
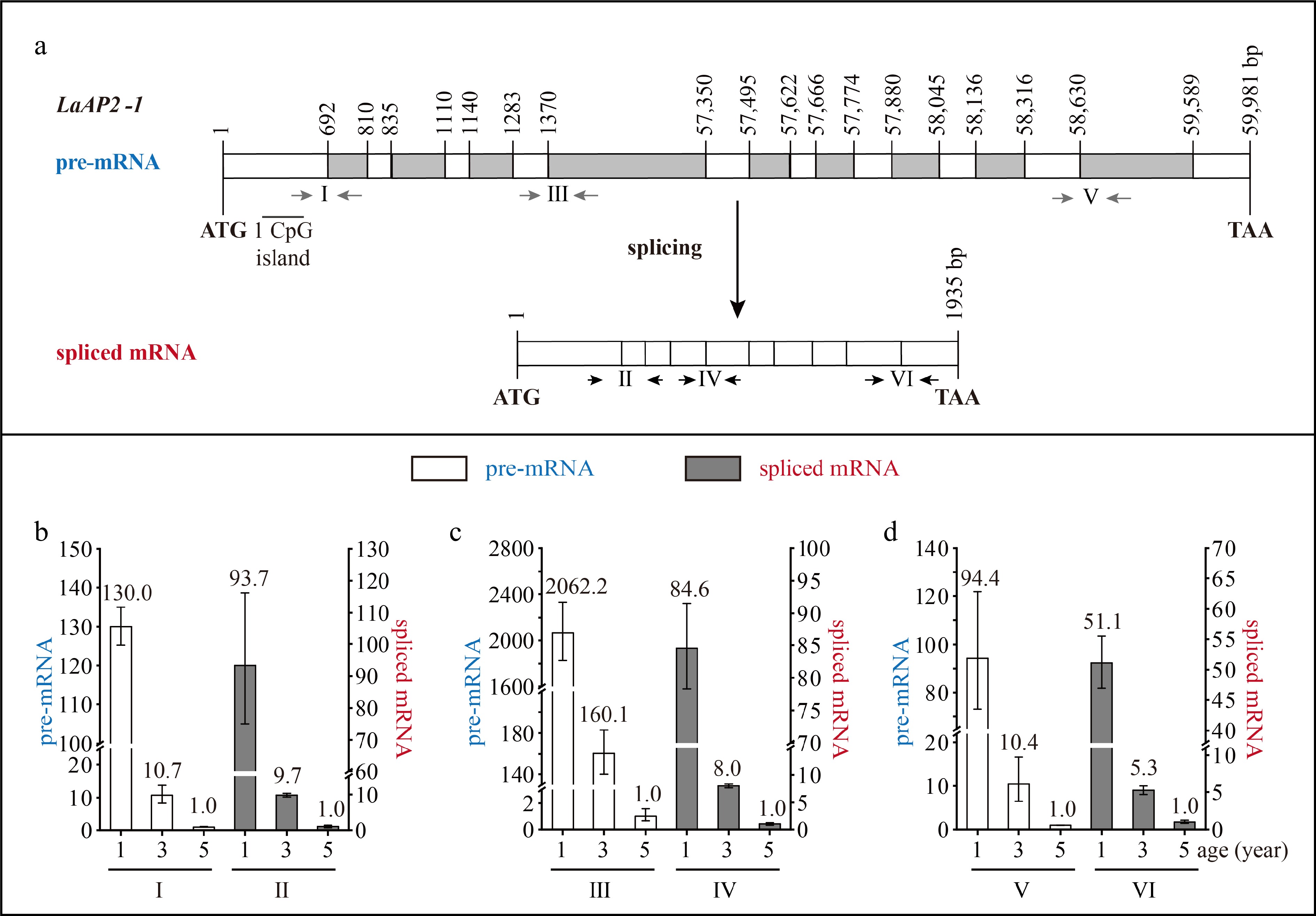
Figure 4.
Structure and expression pattern of LaAP2-1. (a) Schematic representation of the gene structure of LaAP2-1. White indicates exon; gray indicates intron; gray arrows indicate the positions of the primers used to measure the pre-mRNA; and black and horizontal arrows indicate the positions of the primers used to measure the spliced mRNA. After the prediction of DNA methylation, the CpG islands were revealed. (b)–(d) Expression patterns of LaAP2-1 pre-mRNA and spliced mRNA during tree aging detected by three different primer pairs. The lateral branches of 1-, 3-, and 5-year-old active Larix kaempferi trees (n ≥ 6, sampled on 4 July 2019) were used to detect the expression patterns, which were assayed by qRT-PCR with LaEF1A1 as the internal control. The capitalized Roman numerals (I–VI) in panels (a)–(d) represent the different primers.
-
Gene (accession: mRNA/DNA) Primer Sequence (5'-3') Position Size (bp) LaDAL1 (MN790744/WOXR02001943.1) I Forward-CGATGCAGAAGTGGCGCTAA first exon 94 Reverse-GTCAACAGCGCAAAGAAAGGA first intron II Forward-GCTCTCAGTGCTGTGCGAT first exon 249 Reverse-CCGAGATCTTCCCCCAACAAA fourth exon III Forward-AGACCAGATTGAGGAGCTTCG fifth exon 90 Reverse-GGGACGGAATAGCGTGCATTA fifth intron IV Forward-GGTTGAGCTCCTTCAGCGAT third exon 163 Reverse-GCGAAGCTCCTCAATCTGGT fifth exon V Forward-GTACTAACGGGCCTTGGGAT seventh exon 172 Reverse-TTCCAGCTTCAAAAGTGCCAAT seventh intron VI Forward-ACGCAGGTGATGCTAGACCA fifth exon 165 Reverse-CCAAGGCCCGTTAGTACCAG seventh exon LaAGL2-2 (MN790745/WOXR02003181.1) I Forward-GAGTTTGCTAGTGCCGGGTA first exon and first intron 207 Reverse-GTTGGGGGAAGATCTGGGTC first intron II Forward-GGGCTGCTGAAGAAAGCCTA first exon 110 Reverse-TTCATGCCGGCACTAGCAAA second exon III Forward-GTTGGGGGAAGATCTGGGTC fourth exon 242 Reverse-TGCGTTGTGTCGTATTTAGGTC fourth intron IV Forward-TGCAGCAACTCGAACATCAAC fourth exon 76 Reverse-TGGCCTAGCATAACCTGCG fifth exon V Forward-AATTCAAGCCTCCCGACTGT sixth exon 207 Reverse-TCCGGGGACTACATATTGGC sixth intron VI Forward-TGCTCTCTTACACCCGCAAC sixth exon 141 Reverse-CCACCACCCTTGCACGTAT seventh exon LaAP2-1 (MN790757/WOXR02007023.1) I Forward-CCCCGGAGTTCTGAGGAAAC first exon 197 Reverse-TTGCTAGAGGCCTCGTGTTC first intron II Forward-GCTCGCAATATCGTGGAGTG first exon 117 Reverse-TAGCAGCGGCATGAGCAGTA third exon III Forward-CAGCTATCAAGTTTCGAGGCG fourth exon 200 Reverse-ACTGCCATCCAAATGACTACC fourth intron IV Forward-TTCGAGGCGTTGAAGCTGAT fourth exon 131 Reverse-TCCACGAGAGAAACCAGTGC fifth exon V Forward-CTGAAGCTCACATGAGGGAGG ninth exon 158 Reverse-TCCGCTCAGTCCATCTTTATGC ninth intron VI Forward-CCTGACCATCTGGGTAACTGT ninth exon 159 Reverse-TACTGGAGTTGTTGGTCCGC tenth exon Table 1.
Polymerase chain reaction primers for the amplification of age-related transcription factors.
Figures
(4)
Tables
(1)