-
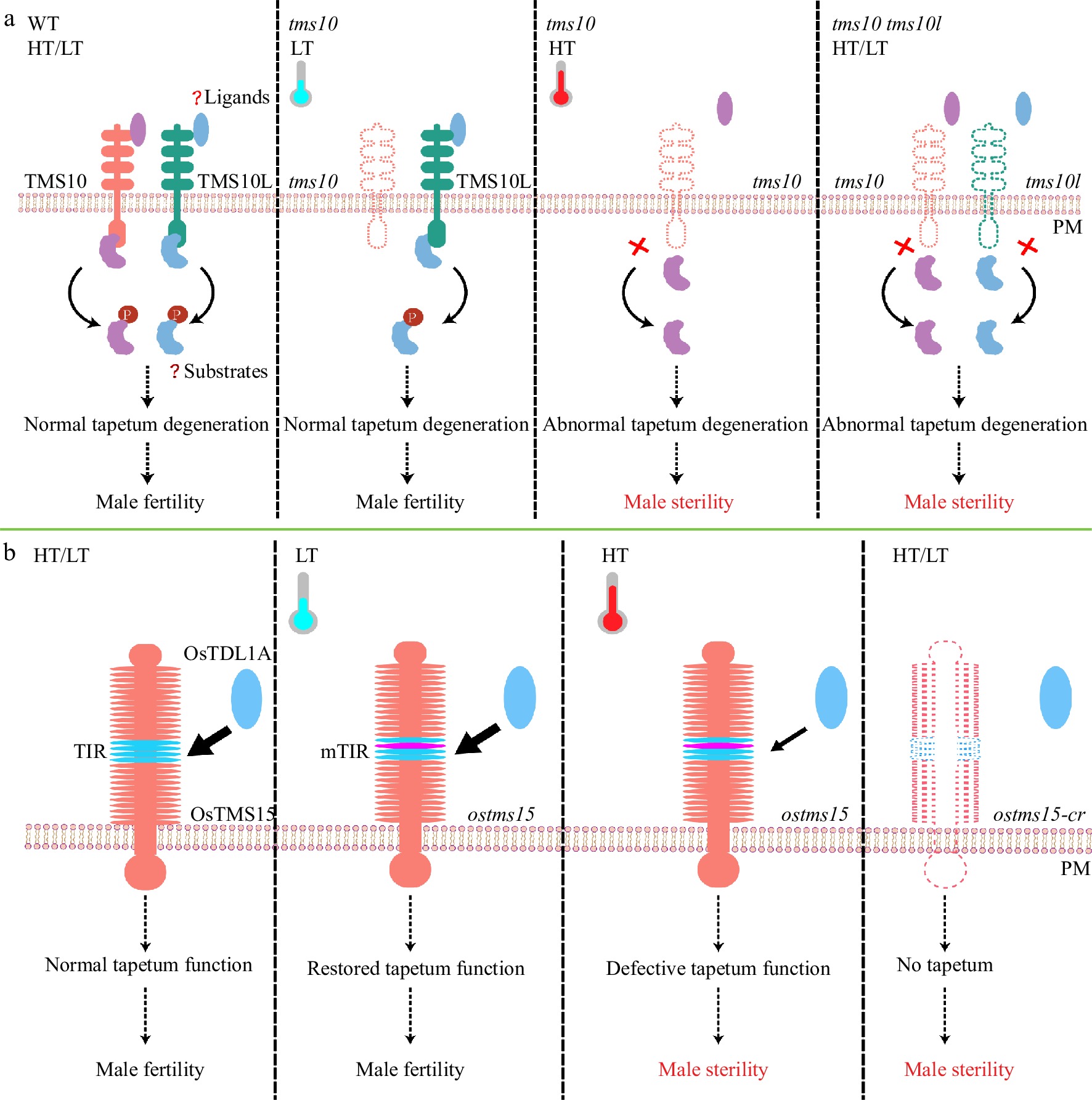
Figure 1.
TMS10/TMS10L, OsTMS5 buffer temperature changes to maintain rice male fertility. (a) In WT, TMS10 and TMS10L relay extracellular signals via substrate phosphorylation (P) to safeguard male fertility at high and low temperatures (HT/LT). In the tms10 mutant at HT, TMS10L is poorly expressed and signals cannot be transduced, resulting in abnormal tapetum degradation and male sterility. At LT, however, TMS10L can restore signal transduction to recover male fertility of the tms10 mutant. The tms10 tms10l double mutant is completely sterile at all temperatures. Red crosses indicate loss of function. (b) In WT, OsTMS15, interacting with its ligand OsTDL1A, initiates tapetum development for pollen formation at HT or LT. In the ostms15 mutant with an amino acid substitution in its TIR motif, the interaction between mTIR and OsTDL1A is reduced, leading to defective tapetum function and male sterility at HT. LT can enhance the interaction and restore tapetum function and male fertility of ostms15 mutant. The knockout line ostms15-cr has no tapetum and is completely sterile at both HT and LT conditions. Os, Oryza sativa (rice).
-
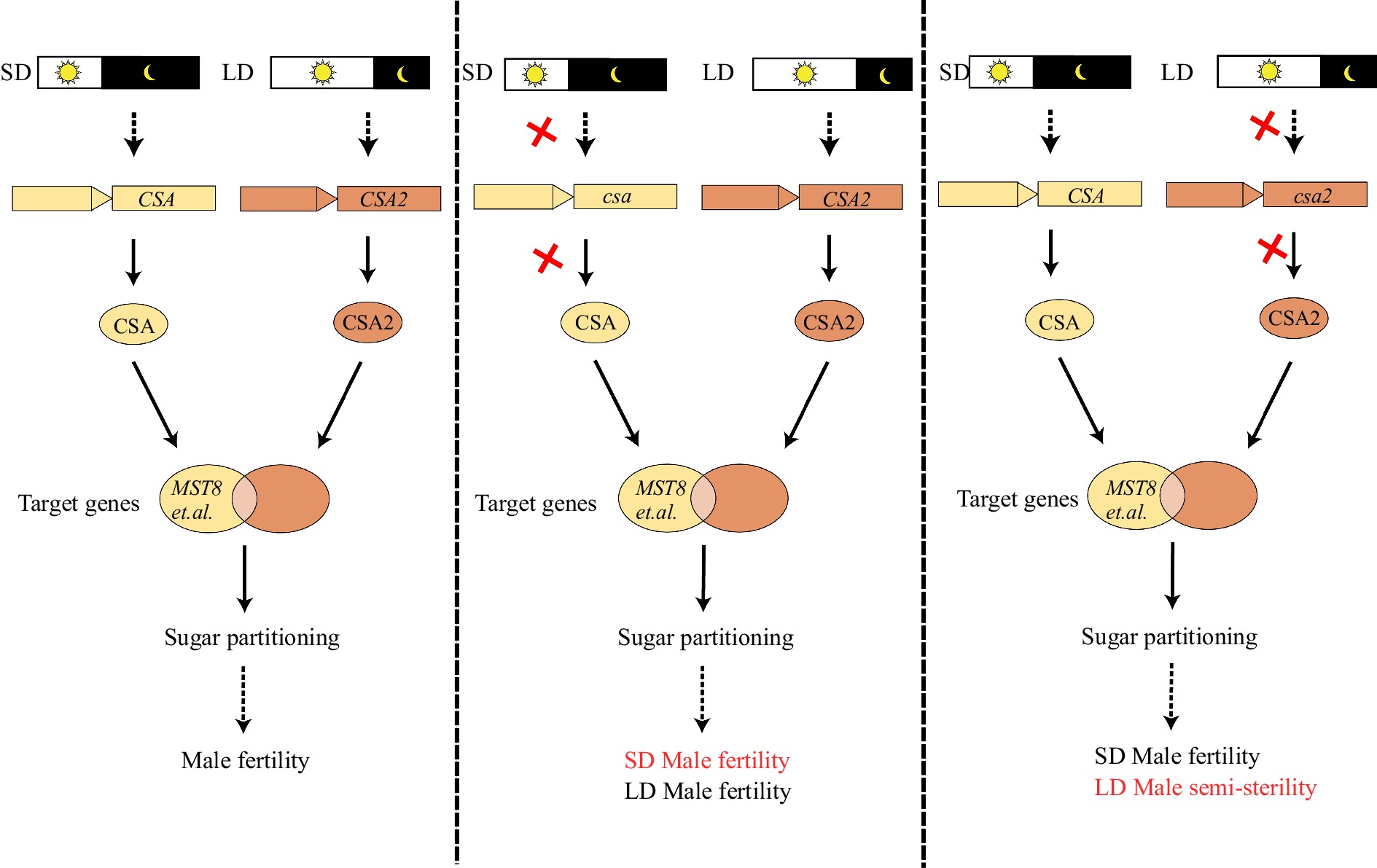
Figure 2.
CSA and CSA2 are a pair of R2R3 MYB transcription factors in rice mediating male fertility adaptation to photoperiod. CSA is a key regulator under short-day (SD) conditions while CSA2 a major regulator under long-day (LD) conditions, both influencing sugar partitioning to the anther. In the csa mutant under SD, downstream target genes, including MST8, are not activated, leading to defective sugar partitioning and male sterility; under LD conditions, CSA2 can restore fertility of the csa mutant. In the csa2 mutant under LD, the loss of CSA2 function reduces activation of downstream target genes, leading to male semi-sterility; while CSA works under SD to restore male fertility. Red crosses indicate loss of function.
-
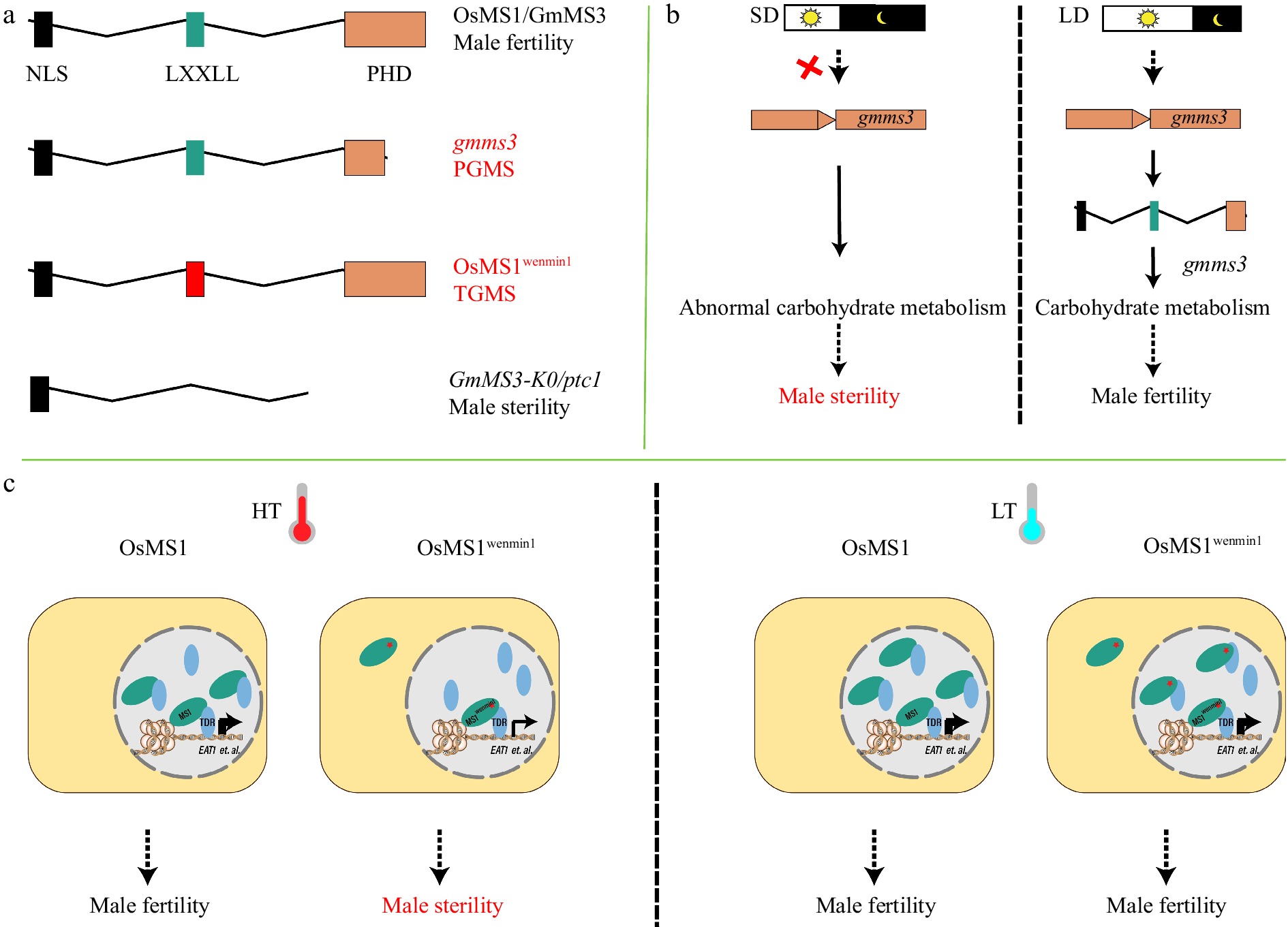
Figure 3.
PHD finger proteins OsMS1 and GmMS3 affect temperature- or photoperiod-sensitive adaption of plant male reproduction in rice and soybean, respectively. (a) Schematic showing domains of plant homeodomain (PHD) finger proteins. Truncation of PHD in gmms3 leads to PGMS in soybean. The L301P substitution in the OsMS1wenmin1 LXXLL (L, leucine; X, any amino acid) motif leads to TGMS in rice. Deletion of the LXXLL and PHD domains in OsMS1/GmMS3 leads to complete male sterility. (b) Short-day (SD) conditions do not induce the expression of gmms3, leading to abnormal carbohydrate metabolism in anthers and male sterility. Long-day (LD) conditions is sufficient to activate gmms3 and restore male fertility. Red crosses indicate loss of function. (c) At high temperature (HT), OsMS1wenmin1 is expressed at low levels and distributed across both cytoplasm and nucleus, so nuclear concentration is insufficient to activate target genes, e.g., EAT1, leading to male sterility. At low temperature (LT), OsMS1wenmin1 is expressed at higher levels and concentrated appropriately in the nucleus to restore male fertility. Os, Oryza sativa (rice); Gm, Glycine max (soybean); NLS, nuclear localization signal.
-
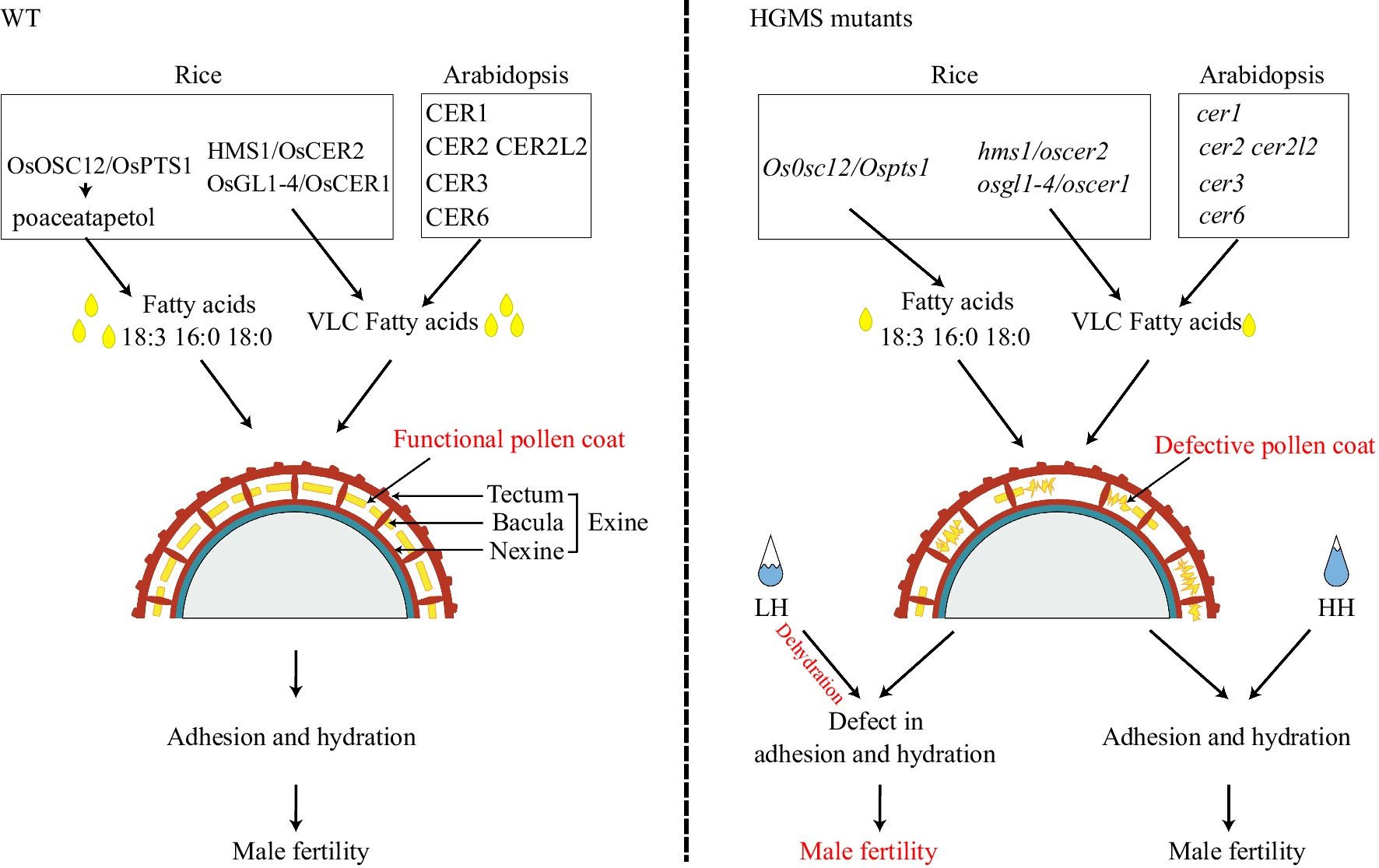
Figure 4.
Pollen coat composition affects fertility at low humidity in rice and Arabidopsis OsOCS12/OsPTS1 influences long-chain fatty acid transport, and OSGL1-4/OsCER1, HMS1/OsCER2, and AtCER1, AtCER2, AtCER2L2, AtCER3, AtCER6 play roles in VLC fatty acid biosynthesis. Mutation in these genes (singly or in pairs) compromises pollen coat composition to reduce adhesion and hydration under low humidity (LH). At, Arabidopsis thaliana; HH, high humidity; VLC, very long chain.
-
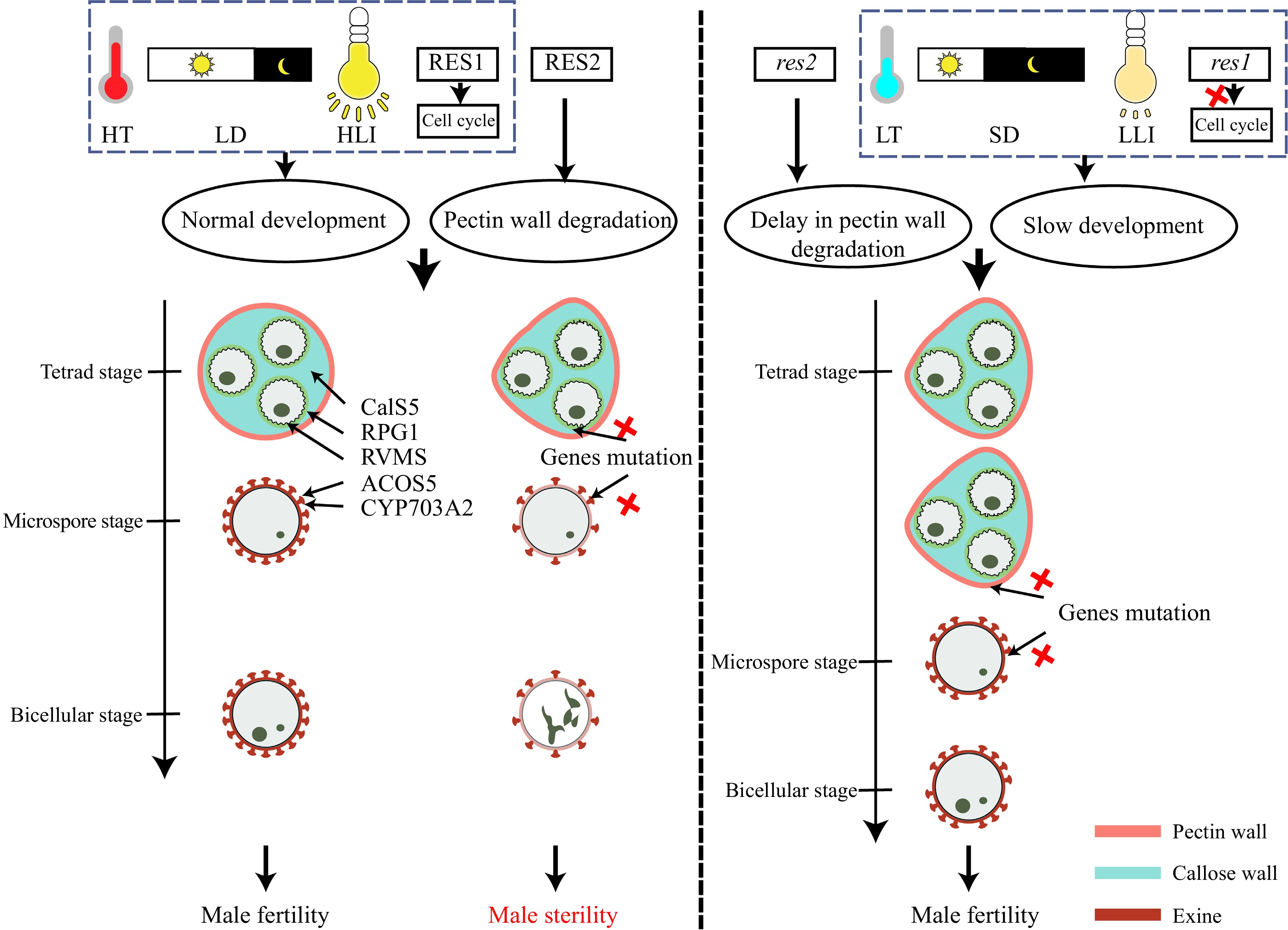
Figure 5.
Slow development and delay in pectin wall degradation can restore fertility in Arabidopsis pollen wall mutants. Low-temperature (LT), short-day (SD), low-light intensity (LLI), and the res1 mutation can slow development of pollen to recover male fertility in Arabidopsis lines defective in pollen wall formation. The res2 mutation delays pectin wall degradation, providing extra protection for developing pollen in pollen-defective mutants. Red crosses indicate loss of function.
-
Species Gene name Gene ID Gene product Type of
EGMSPathway Reference Arabidopsis ACOS5 At1g62940 acyl-CoA synthetase 5 P/TGMS Pollen exine formation [36,38] RVMS At4g10950 GDSL lipase P/TGMS Pollen nexine formation [36,38] CalS5 At2g13680 callose synthase 5 P/TGMS Pollen exine patterning [36,38] RPG1/SWEET8 At5g40260 SWEET8 TGMS Pollen nexine formation [34,35,38,
111,112]CYP703A2 At1g01280 cytochrome P450 703A2 P/TGMS Pollen exine formation [36,38] ABCG26 At3g13220 ATP-binding cassette transporter G26 TGMS Pollen exine formation [38] TMS1 At3g08970 HSP40 TGMS Growth of pollen tubes, unfolded protein response [113,114] NPU At3g51610 ATP-dependent helicase/deoxyribonuclease subunit B P/TGMS Pollen primexine
deposition[36] IRE1A IRE1B At2g17520 At5g24360 IRE TGMS Pollen coat formation, unfolded protein response [115] AtSec62 At3g20920 Translocation protein TGMS Protein translocation and
secretion[116] PEAMT At3g18000 S-adenosyl-l-methionine: phosphoethanolamine N-methyltransferase TGMS Signal transduction [117] AtPUB4 At2g23140 E3 ligase TGMS Protein degeneration [118] COI1 LOC9315901 F box protein TGMS Protein degeneration [119] ICE1 At3g26744 MYC-like bHLH
transcriptional activatorHGMS Anther dehiscence, transcriptional regulation [120] MYB33 MYB65 At5g06100 At3g11440 R2R3 MYB transcription
factorTGMS Tapetum PCD, transcriptional regulation [62] CER1 At1g02205 Acyl-CoA synthetase HGMS Pollen coat function [82] CER3 At5g57800 Acyl-CoA synthetase HGMS Pollen coat function [84] CER6/CUT1 At1g68530 Acyl-CoA synthetase HGMS Pollen coat function [89] CER8, LACS4 At2g47240, At4g23850 Acyl-CoA synthetase HGMS Pollen coat function [90] FKP At4g11820 3-hydroxy-3-methylglutaryl-coenzyme A Synthase HGMS Pollen coat function [121] Rice UGP1 Os09g0553200 UDP-glucose pyrophosphorylase1 TGMS RNA processing [27] TMS5 Os02g0214300 RNase ZS1 TGMS RNA processing [22] TMS10 -TMS10L Os02g0283800-LOC_Os03g49620 LRR-RLK TGMS Signaling transduction [19] TMS9-1/OSMS1 Os09g0449000 PHD finger protein TGMS Protein location and transcriptional regulation [63] AGO1d Os06g0729300 Argonaute protein TGMS PhasiRNAs production [53,54] HMS1-HMS1I Os03g0220100-Os01g0150000 3-ketoacyl-CoA synthase 6 very-long-chain enoyl-CoA reductase HGMS Pollen coat function [32] OSOSC12 /OSPTS1 Os08g0223900 Bicyclic triterpene synthase HGMS Pollen coat function [81] OsCER1/ OsGL1-4 Os02g0621300 Acyl-CoA synthetase HGMS Pollen coat function [91] CSA Os01g0274800 R2R3 MYB transcription
factorrPGMS Sugar distribution, transcriptional regulation [23,24] CSA2 Os05g049060 R2R3 MYB transcription
factorPGMS Sugar distribution, transcriptional regulation [26] PMS1 AK242308 lncRNA PGMS lncRNA regulation [29] P/TMS12-1 (PMS3) Os12g0545900 lncRNA, smR5864 P/TGMS lncRNA regulation and smRNA regulation [51,52] OsPDCD5 AY327105 Programmed cell death 5 protein PGMS Tapetum PCD [122] OSMYOXIB Os02g0816900 Myosin XI B PGMS Nutrition transport, protein location [123] OsNP1/OsTMS18 Os10g0524500 Glucose–methanol–choline oxidoreductase TGMS Pollen exine formation [97] ORMDL/tms2 Os07g0452500 Orosomucoid TGMS Sphingolipid homeostasis,PCD [124] OsOAT LOC_Os03g44150 Ornithine δ-aminotransferase rTGMS Cold tolerance [125] HSP60-3B LOC_Os10g32550 Heat Shock Protein60-3B TGMS Starch granule biogenesis, reactive oxygen species (ROS) levels [126] OsTMS15 LOC_Os01g68870 LRR-RLK TGMS Tapetum development [44] OsAL5 LOC_Os05g34640 Alfin like TGMS TMS5 expression [60] Maize TMS5 LOC100285786 RNase ZS1 TGMS mRNA decay [127] DCL5 LOC103643440 Dicer-like 5 TGMS PhasiRNAs production [56] MAGO1, MAGO2 Zm00001d007786, Zm00001d013063 MALE-ASSOCIATED ARGONAUTE TGMS Pre-meiotic phasiRNA pathways [55] INVAN6 Zm00001d015094 Alkaline/neutral invertase TGMS Sugar accumulation, metabolism, and signaling [128] Brassica
napusBnChimera BnRfb naA7.mtHSP70-1-like rTGMS Fatty acid synthesis [99] Barley HvMS1 LOC123121697 PHD finger protein rTGMS Transcriptional regulation [129] Soybean MS3 GLYMA_02G107600 PHD finger protein rPGMS Transcriptional regulation [64] EGMS, environment-sensitive genic male sterility; PGMS, photoperiod-sensitive genic male sterility; rPGMS, reverse photoperiod-sensitive genic male sterility; TGMS, thermo-sensitive genic male sterility; rTGMS, reverse thermo-sensitive genic male sterility; HGMS, humidity-sensitive genic male sterility. Table 1.
Genes involved in environmental adaption of plant male reproduction.
Figures
(5)
Tables
(1)