-
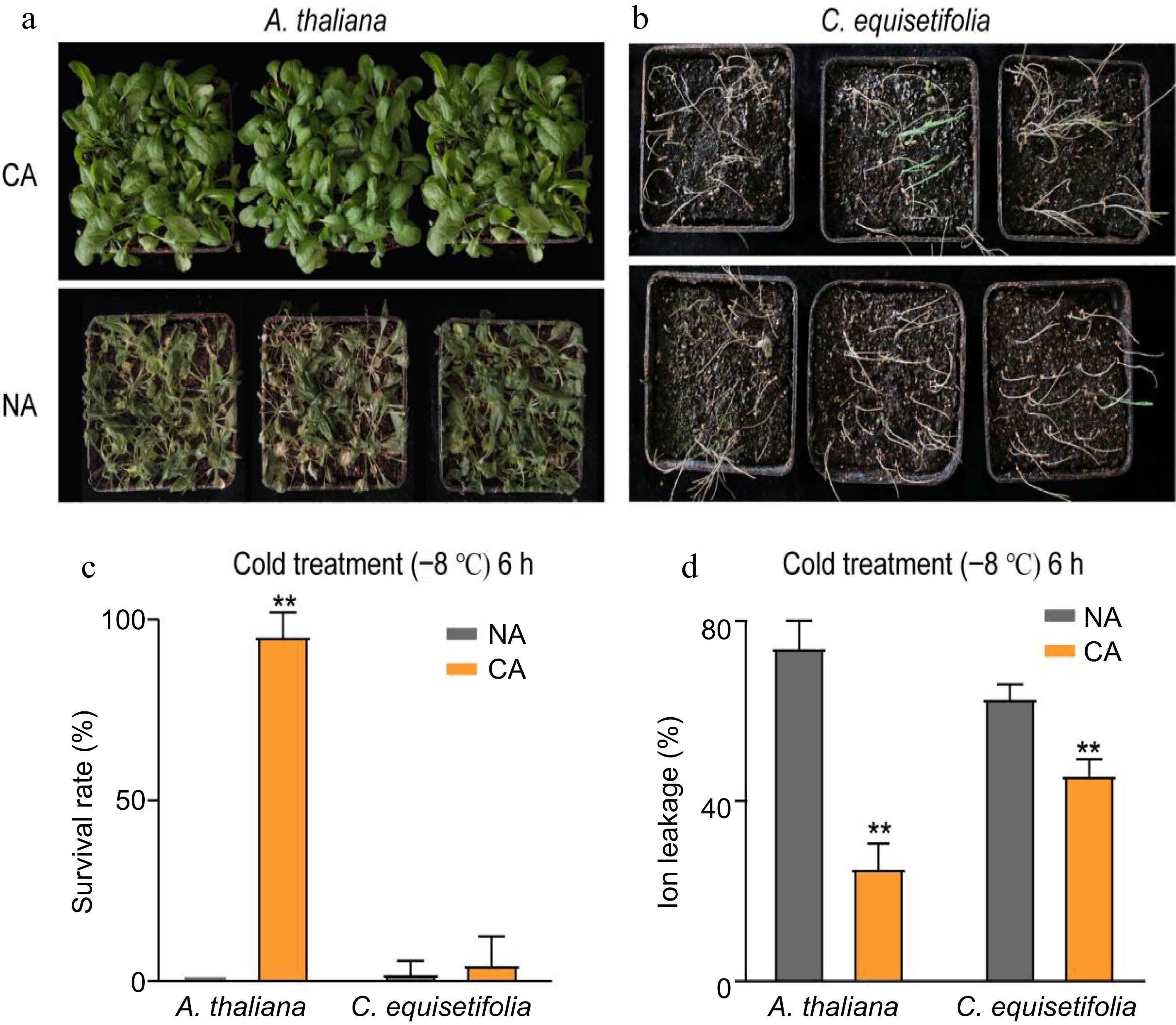
Figure 1.
Analysis of cold resistance of A. thaliana and C. equisetifolia. (a), (b) Freezing phenotypes (c) survival rates, and (d) ion leakage, of three-week-old wild type A. thaliana and two-month-old C. equisetifolia. The seedlings were grown under a 16-h-white light/8-h-dark photoperiod (LD) at 25 °C and either directly exposed to freezing treatment (non-acclimated, NA), or treated at 4 °C for 4 d or 7 d before freezing treatment (cold-acclimated, CA). Representative photographs were taken after a 3-d recovery period, survival rates were calculated, and the ion leakage was measured. Data are presented as the mean ± standard deviation of mean of three biological replicates. Asterisks represent significant differences compared to the non-acclimated plant. ** p < 0.01.
-
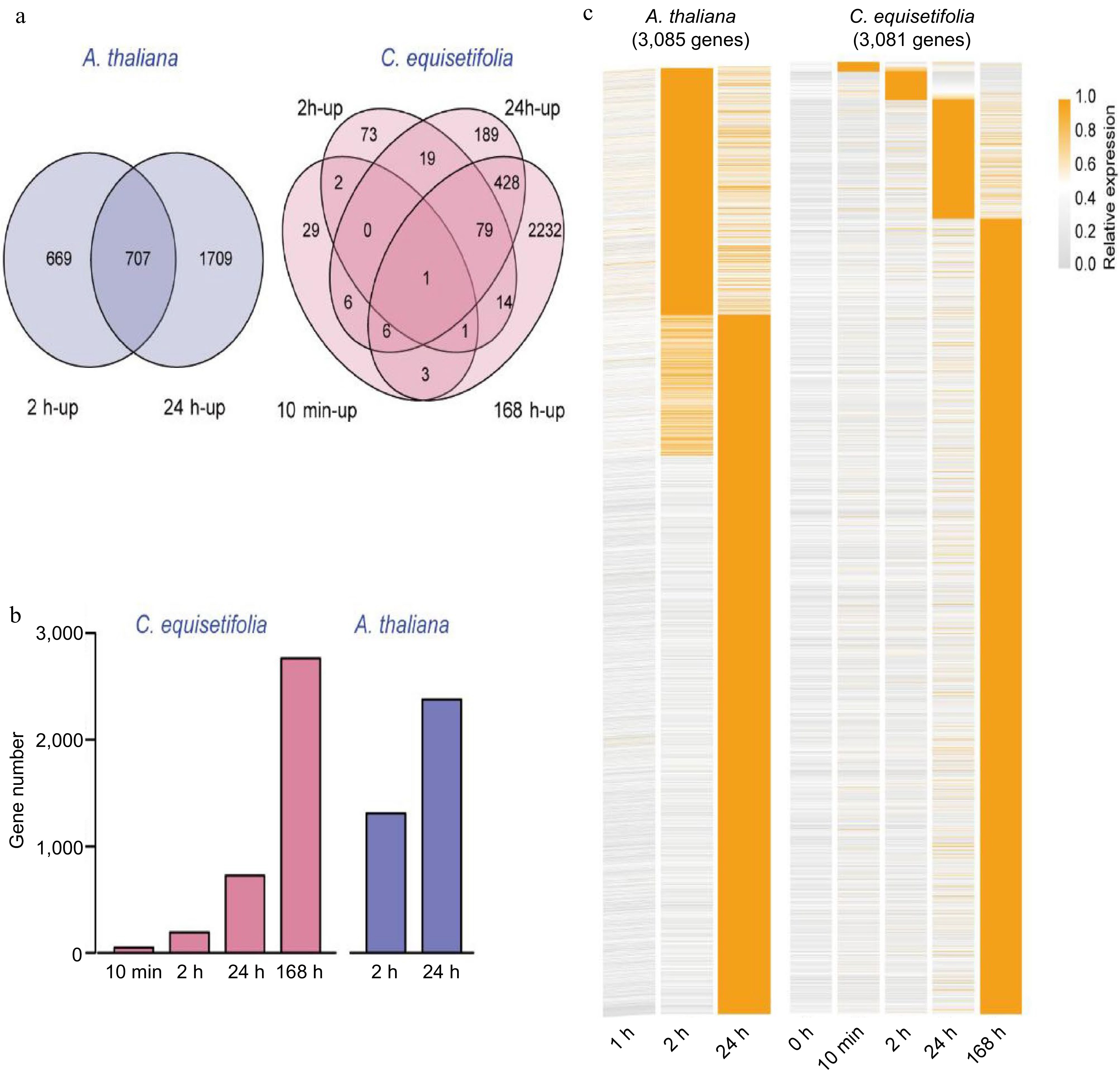
Figure 2.
Comparison of the number of cold-induced genes at different time points in A. thaliana and C. equisetifolia. (a) Venn diagram illustrating the cold-induced genes at 10 min, 2, 24 and 168 h (C. equisetifolia) and 2 h, 24 h (A. thaliana) after chilling treatment. (b) The number of cold-induced genes between the different plants and time points. (c) Expression profiles and clustering of 3,081 cold-induced genes in C. equisetifolia and 3,085 cold-induced genes in A. thaliana at different times after cold acclimation.
-
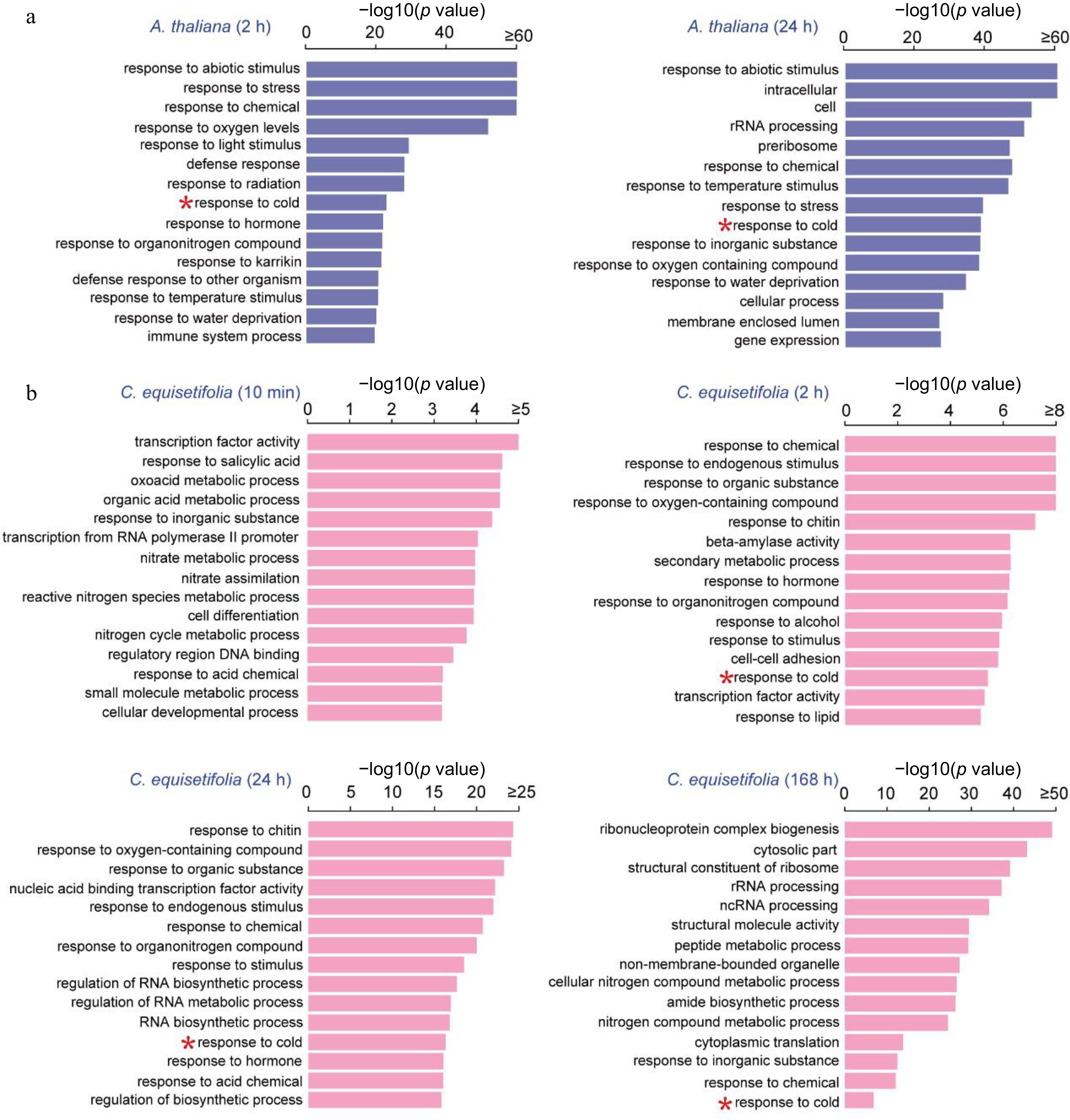
Figure 3.
GO enrichment analysis of cold-induced genes at different time points in A. thaliana and C. equisetifolia under 4 °C cold treatment. (a) A. thaliana, (b) C. equisetifolia.
-
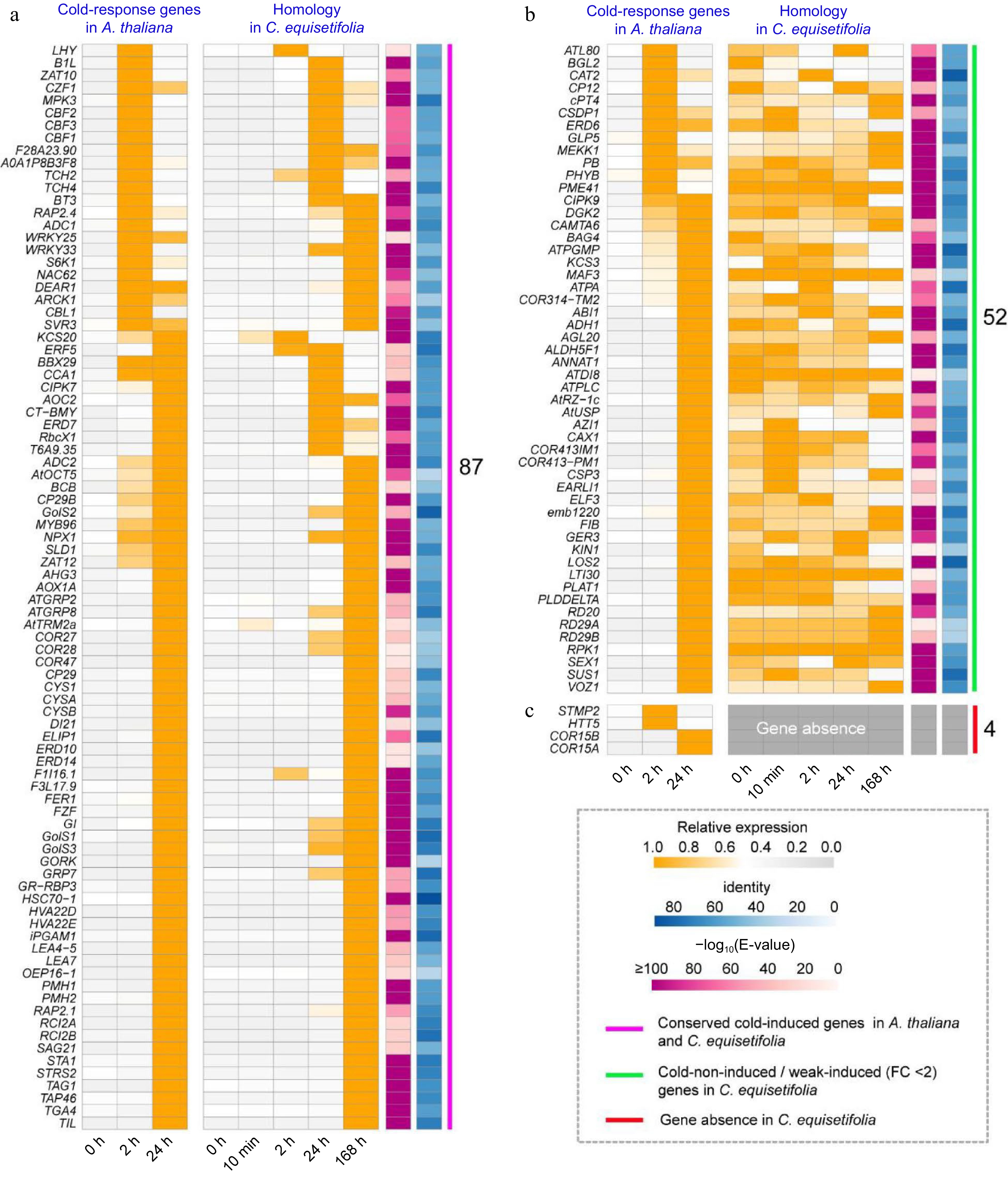
Figure 4.
Comparison of the 'cold response entries' in A. thaliana and C. equisetifolia. (a) Conserved cold-induced genes in A. thaliana and C. equisetifolia. (b) Unresponsive to cold stress/ weakly cold-induced (FC < 2) genes in C. equisetifolia. (c) Gene absent in C. equisetifolia.
-
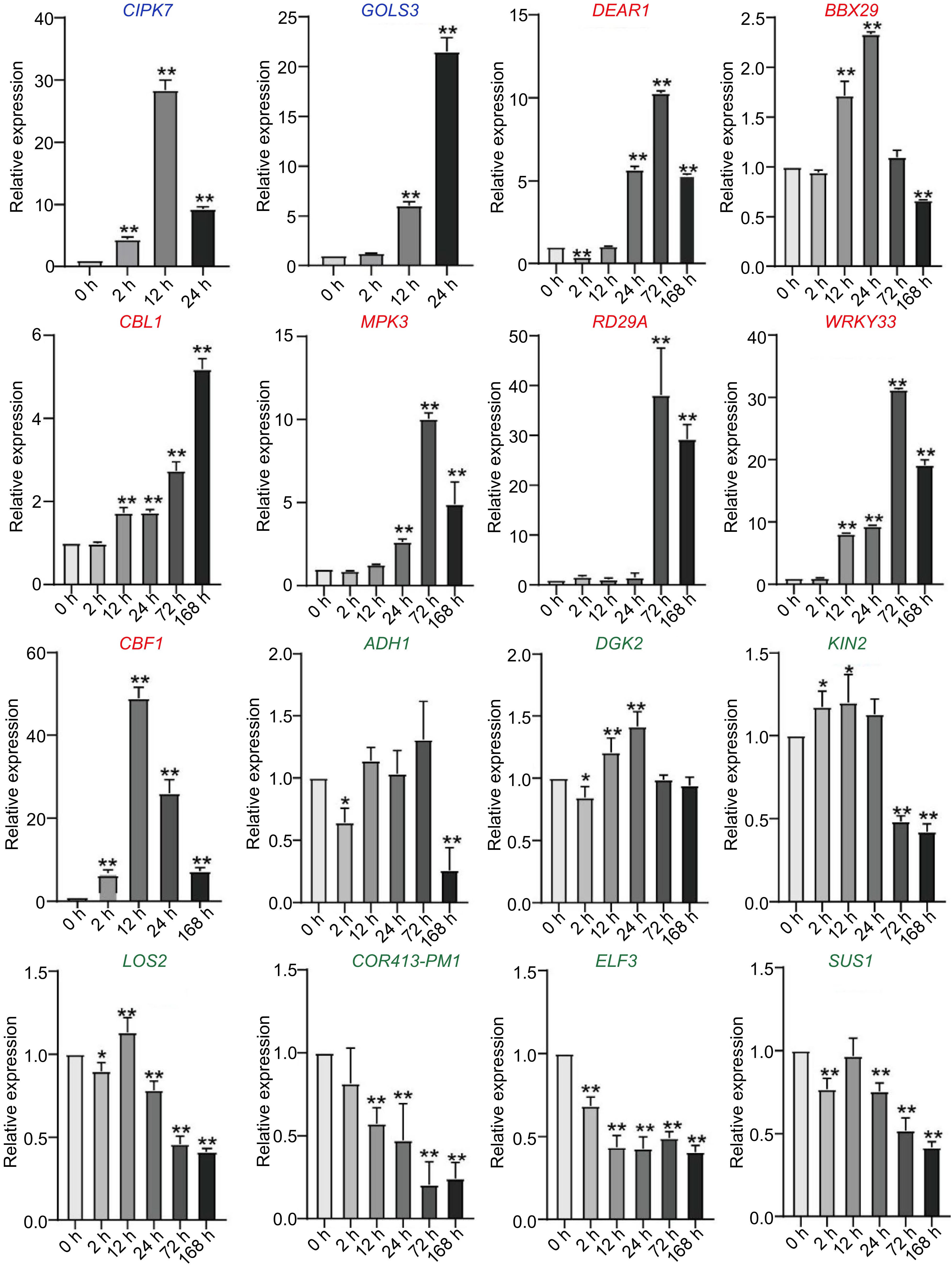
Figure 5.
Expression of cold-response genes in C. equisetifolia, containing two similar expression pattern genes (CIPK7, GOLS3), seven delayed genes (DEAR1, BBX29, CBL, MPK3, RD29A, WRKY33, CBF1), four weakly responsive genes (ADH1, DGK2, KIN1, LOS2) and three non-responsive genes (COR413-PM1, ELF3, SUS1). CIPK7, CBL-interacting protein kinase 7; GOLS3, galactinol synthase 3; DEAR1, cooperatively regulated by ethylene and jasmonate 1; BBX29, B-box type zinc finger family protein; CBL, calcineurin B-like protein 1; MPK3, mitogen-activated protein kinase 3; RD29A, low-temperature-responsive protein 78 (LTI78); WRKY33, WRKY DNA-binding protein 33; ADH1, alcohol dehydrogenase 1; DGK2, diacylglycerol kinase 2; KIN1, stress-responsive protein (KIN1)/stress-induced protein (KIN1); LOS2, involved in light-dependent cold tolerance and encodes an enolase; COR413-PM1, cold regulated 413 plasma membrane1; ELF3, hydroxyproline-rich glycoprotein family protein; SUS1, encodes a protein with sucrose synthase activity. The values are the mean ± standard deviation of three biological replicates. Relative expression in untreated plants (0 h) was set to 1. *p < 0.05, **p < 0.01.
-
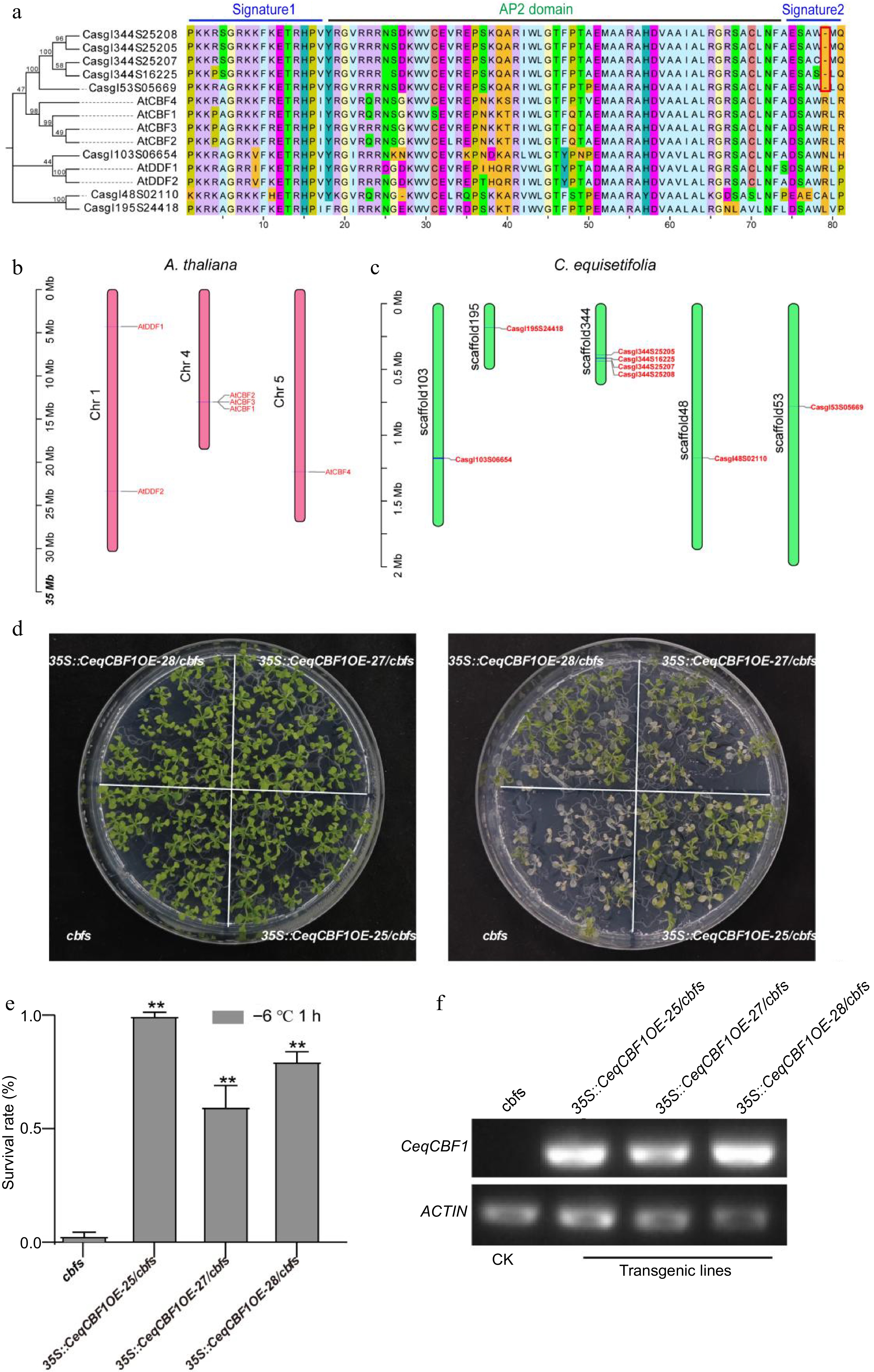
Figure 6.
CBF in C. equisetifolia was involved in cold regulation. (a) Alignment of AP2/ERE domain amino acid sequences between A. thaliana and C. equisetifolia. (b), (c) Chromosome positions of CBF genes. (d) Freezing phenotypes, (e) survival rates and (f) the expression of CeqCBF1 in A. thaliana cbf triple mutants and transgenic lines with overexpressing of CeqCBF1. Two-week-old plants grown on MS plates at 22 °C were treated at −6 °C for 1 h after cold acclimation at 4 °C for 3 d. Asterisks indicate significant differences compared to the cbfs mutant plants. **p < 0.01.
-
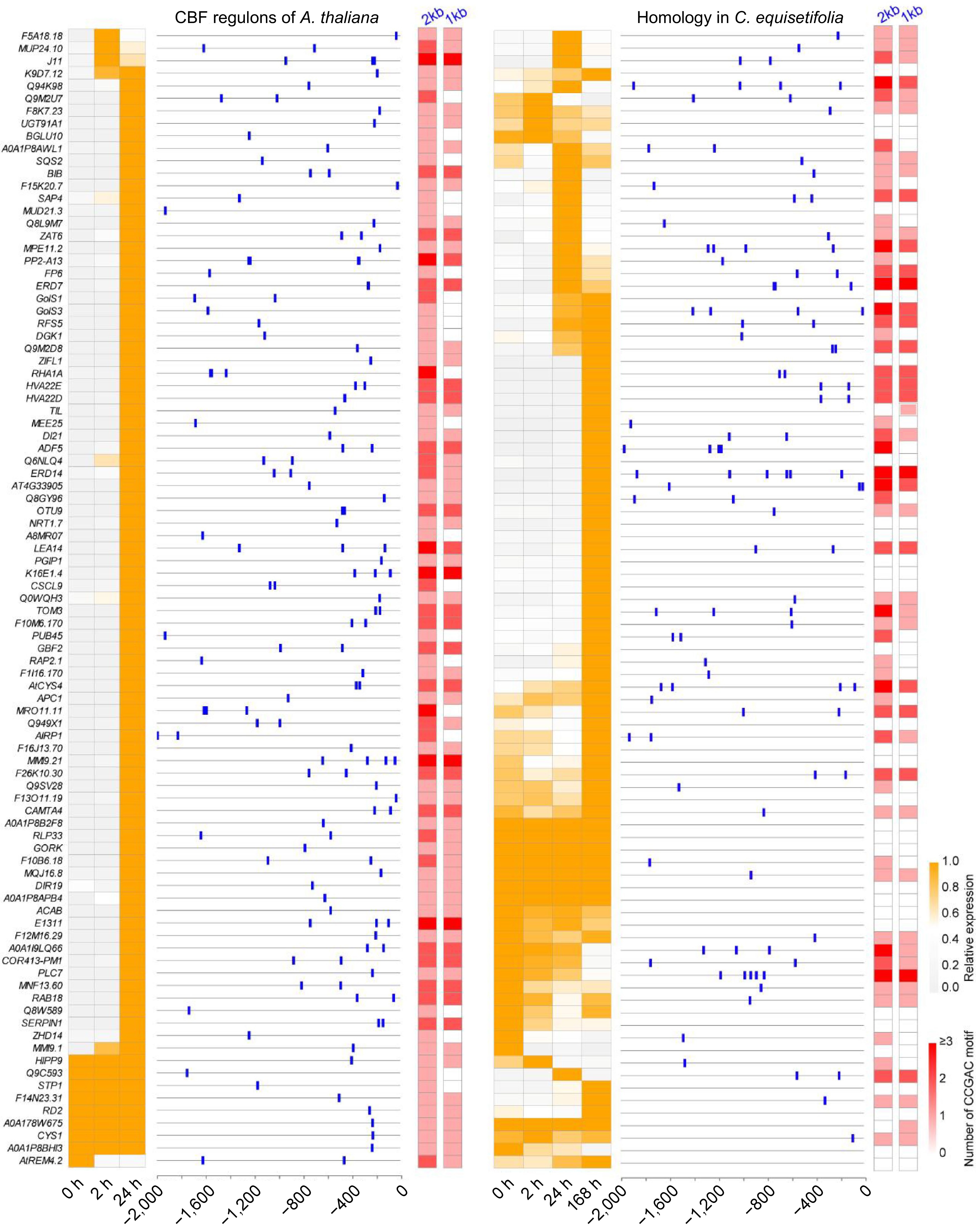
Figure 7.
Heatmap and CRT/DRE element analysis of downstream cold response genes directly regulated by CBF in A. thaliana and C. equisetifolia.
-
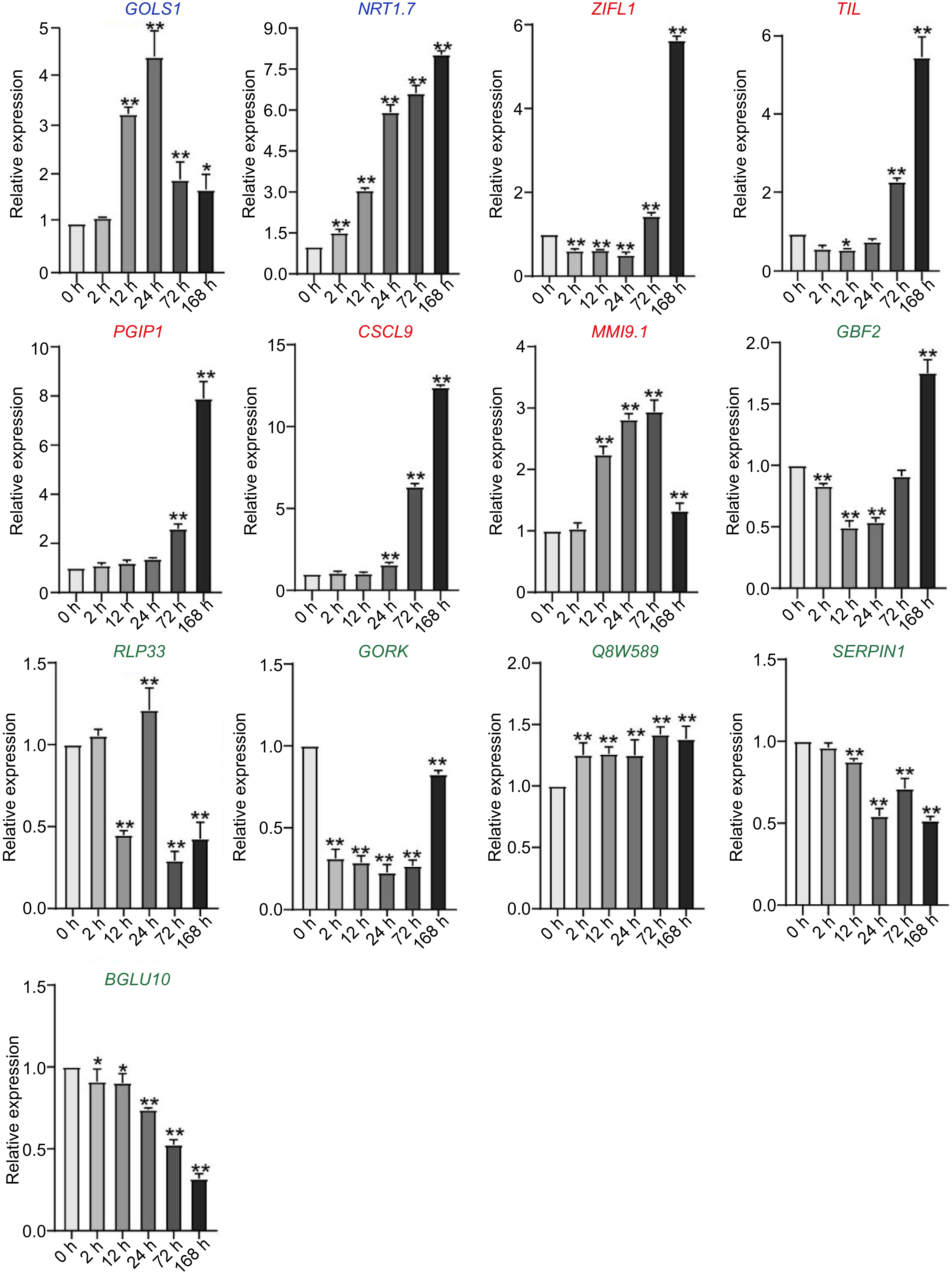
Figure 8.
The expression of downstream genes without DRE elements but directly regulated by CBF in C. equisetifolia, containing two similar expression pattern genes (GOLS1, NRT1.7), five delayed genes (ZIFL1, TIL, PGIP1, CSCL9, MMI9.1) and six weakly responsive and non-responsive genes (GBF2, RLP33,GORK,Q8W589, SERPIN1, BGLU10), GOLS1, galactinol synthase 1; NRT1.7, NITRATE TRANSPORTER 1.7; ZIFL1, ZINC INDUCED FACILITATOR-LIKE 1; TIL, temperature-induced lipocalin; PGIP1, POLYGALACTURONASE INHIBITING PROTEIN 1; GBF2, G-box binding factor 2; RLP33, receptor like protein 33; GORK, GATED OUTWARDLY-RECTIFYING K+ CHANNEL; SERPIN1, Inhibitor of pro-apoptotic proteases, which is involved in the regulation of the programmed cell death induction; BGLU10, BETA GLUCOSIDASE 10. The values are the mean ± standard deviation of three biological replicates. Relative expression in untreated plants (0 h) was set to 1. *p < 0.05, **p < 0.01.
-
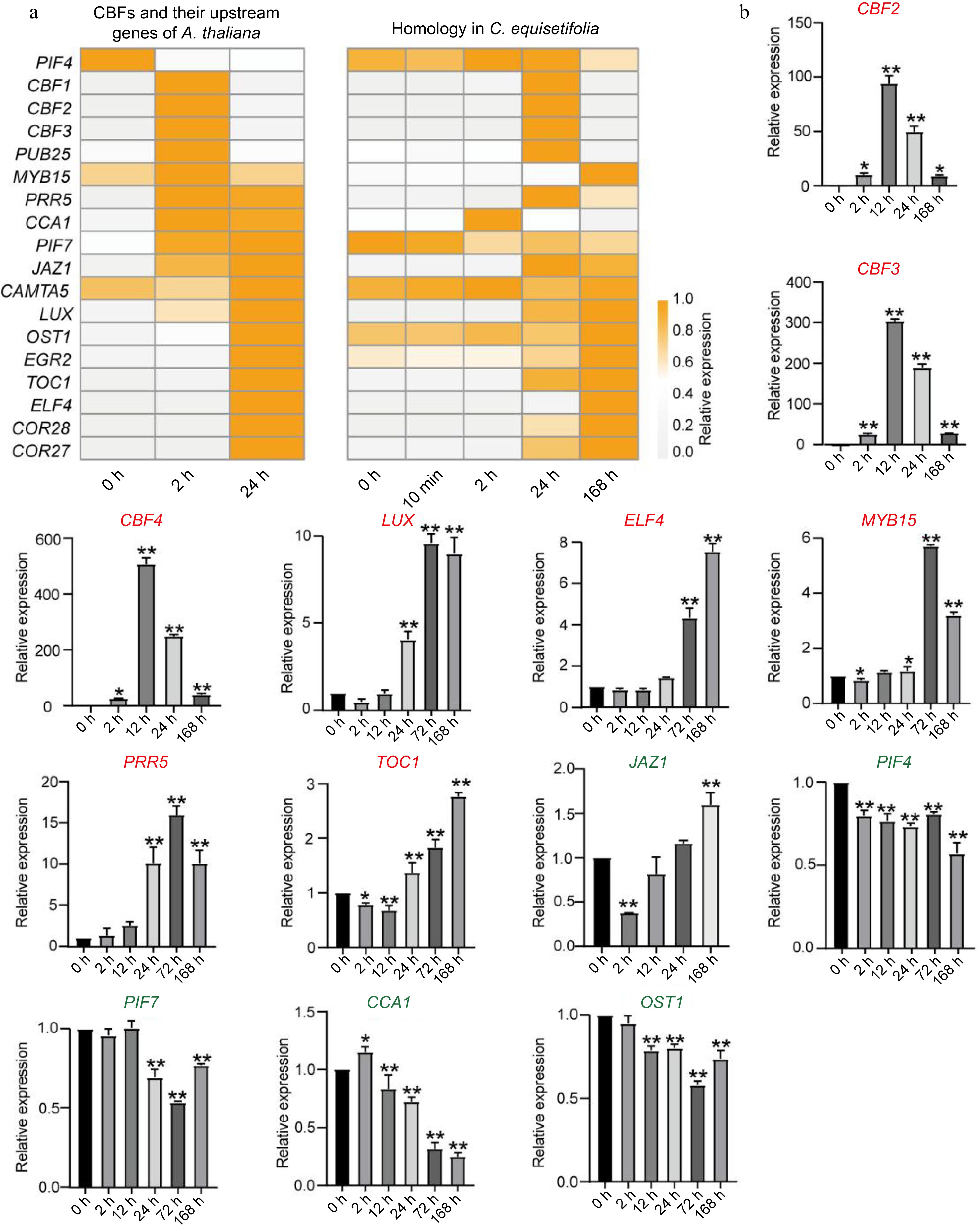
Figure 9.
Comparison of upstream regulatory genes for CBF in A. thaliana and C. equisetifolia. (a) Heatmap of CBF upstream regulatory genes in A. thaliana and C. equisetifolia. (b) The expression of CBF upstream gene in C. equisetifolia. CBF, C-repeat/DRE binding factor; LUX, Homeodomain-like superfamily protein; ELF4, early flowering-like protein; MYB15, myb domain protein 15; PRR5, two-component response regulator-like protein; TOC1, CCT motif -containing response regulator protein; JAZ1, jasmonate-zim-domain protein 1; PIF4/7, phytochrome-interacting factor4/7; CCA1, circadian clock associated 1; OST1, Protein kinase superfamily protein. Relative expression in untreated plants (0 h) was set to 1. *p < 0.05, **p < 0.01.
-
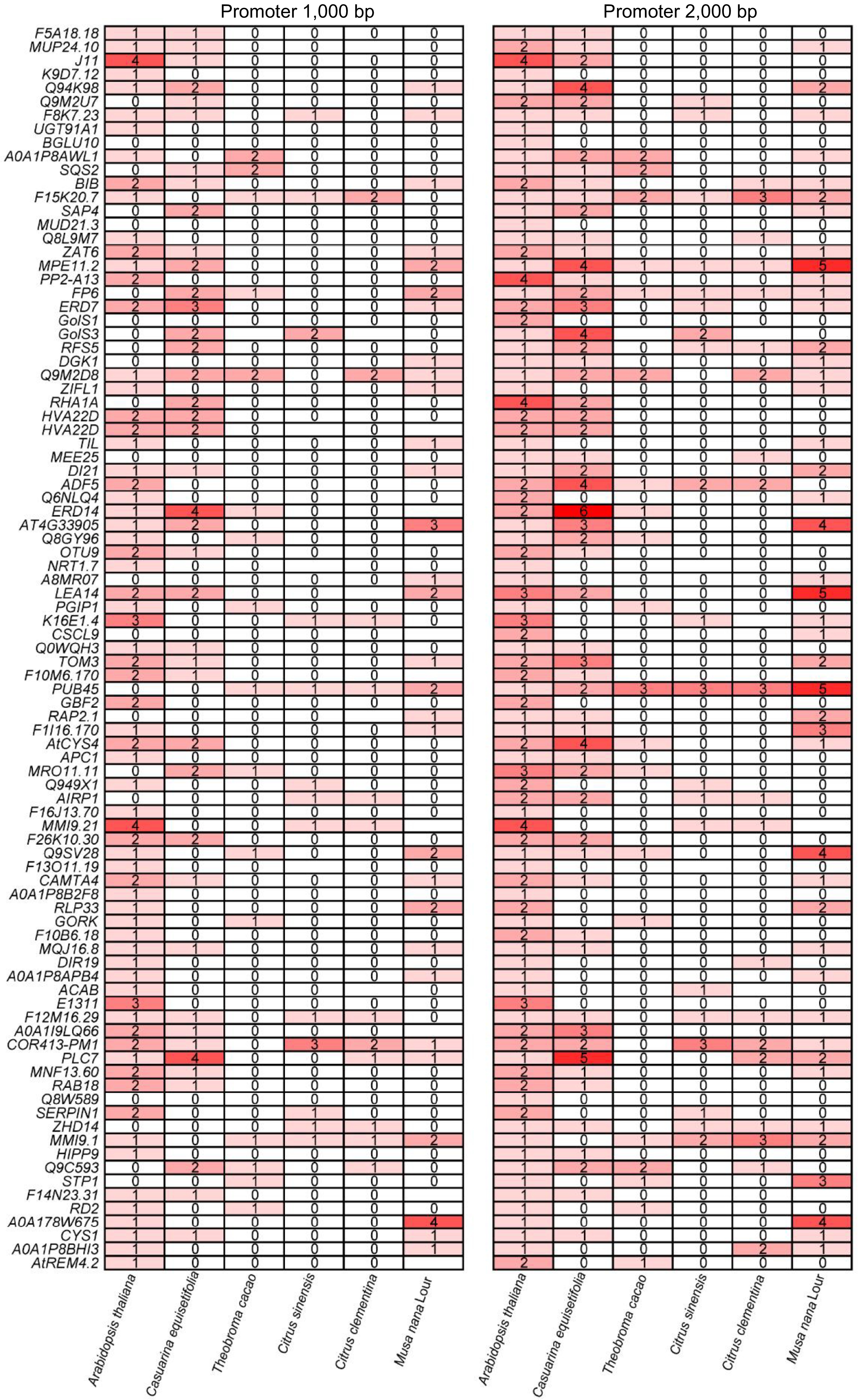
Figure 10.
CRT/DRE element analysis of downstream cold response genes directly regulated by CBF in other tropical and subtropical plants.
-
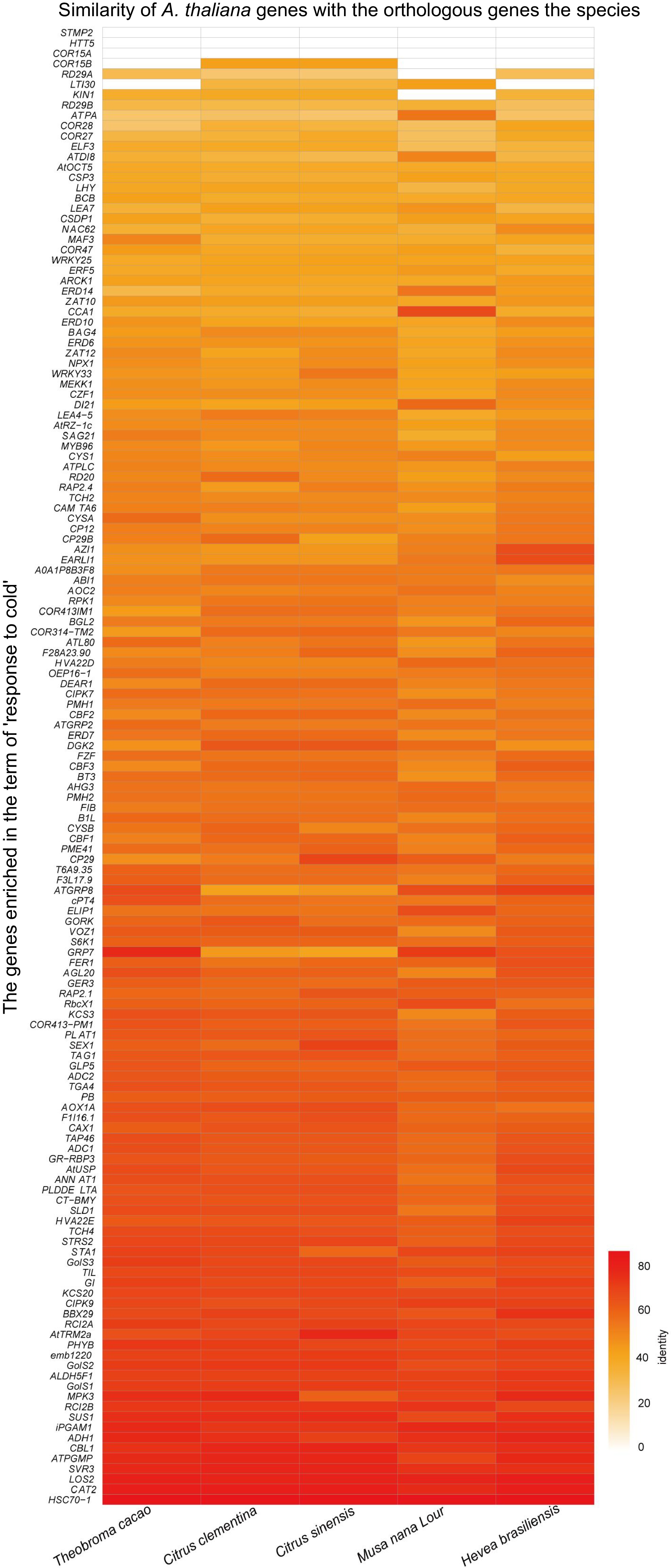
Figure 11.
Homologous cold response genes in A. thaliana and other tropical/subtropical plants.
-
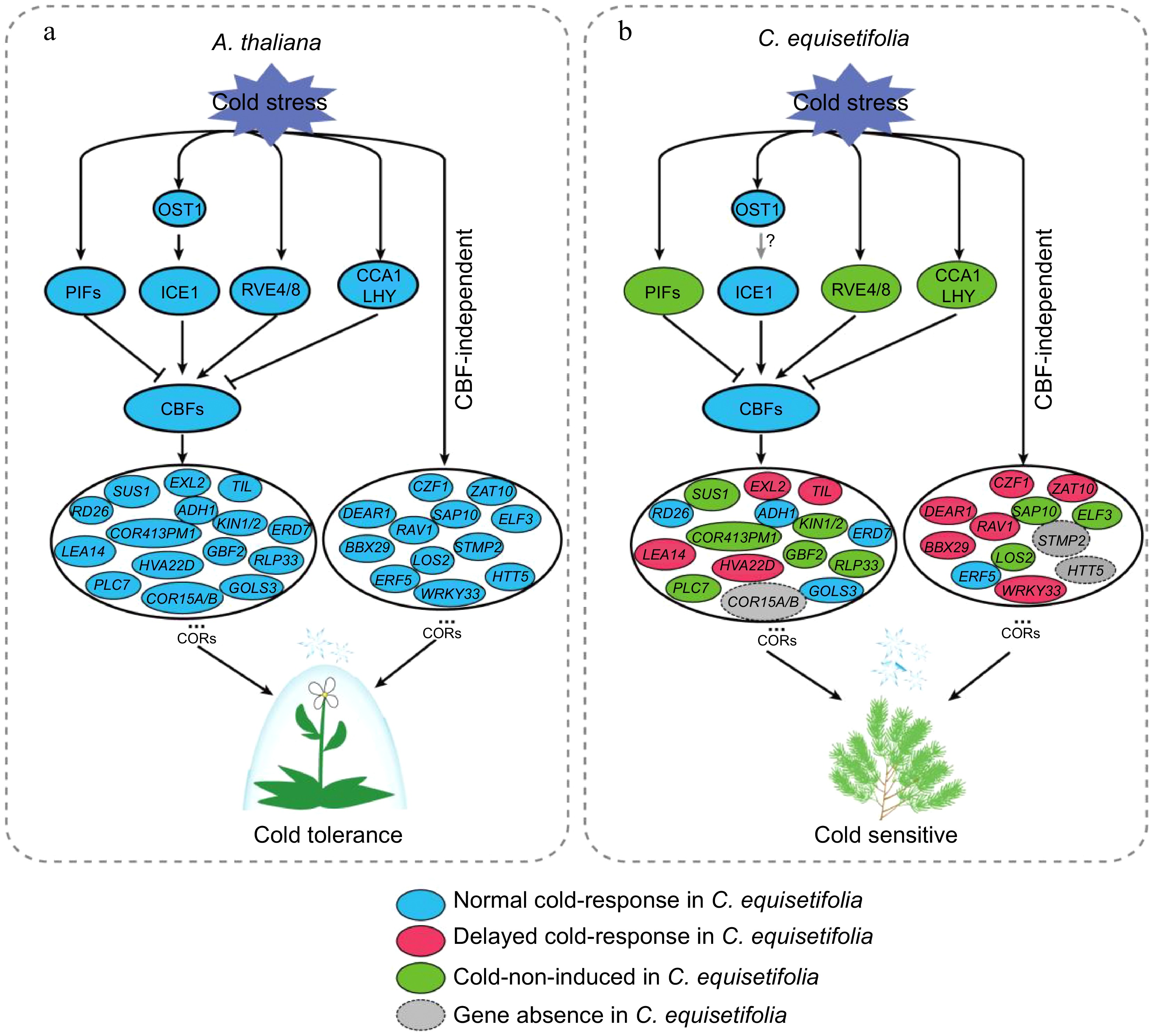
Figure 12.
Comparative pattern plot of cold-response regulatory pathway in A. thaliana and C. equisetifolia.
Figures
(12)
Tables
(0)