-
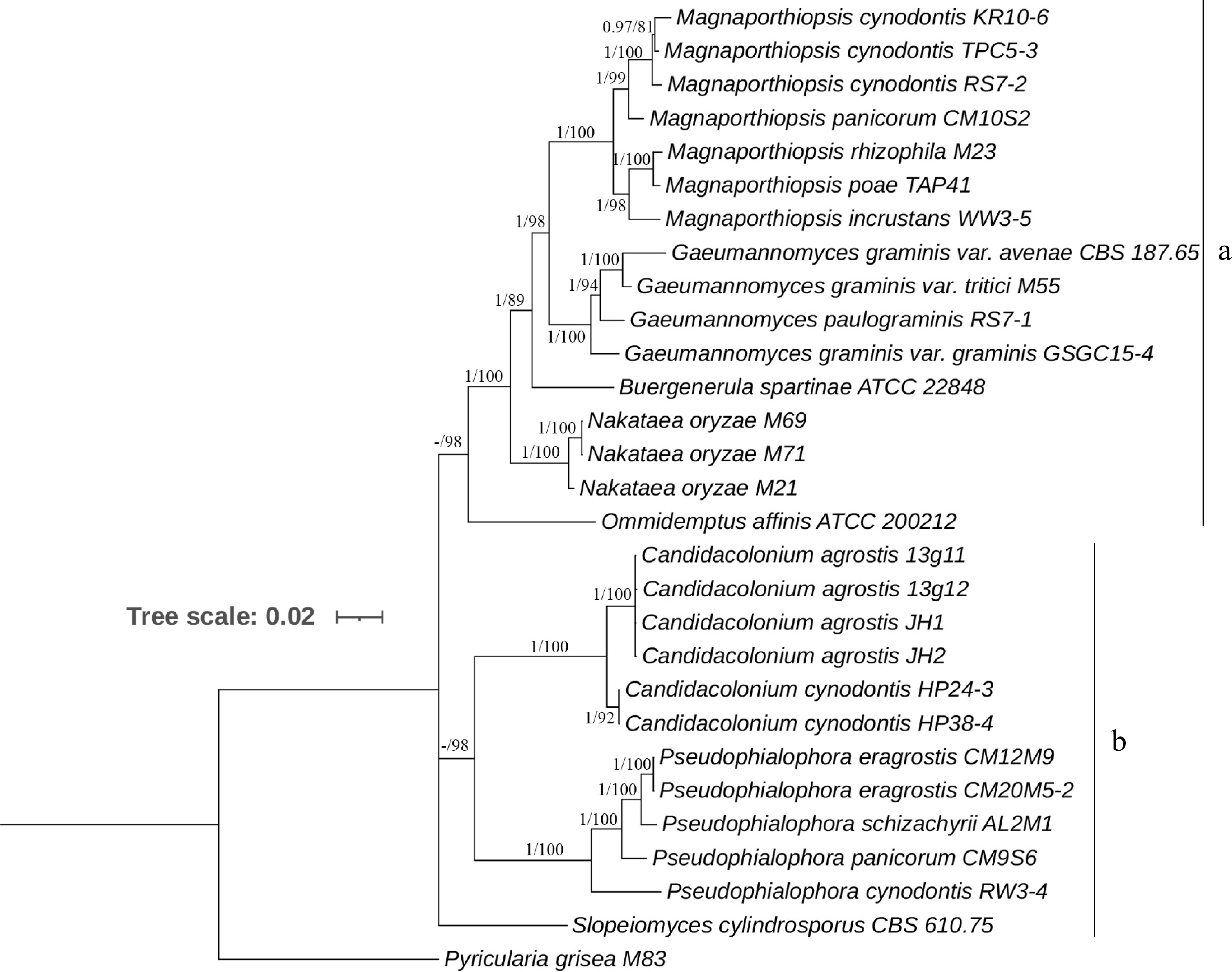
Figure 1.
Majority rule Bayesian phylogenetic tree derived from the combined ITS, LSU, SSU, MCM7, RPB1, and TEF1 DNA sequence data sets. Nodes having Bayesian inference posterior probabilities ≥ 0.95 and maximum likelihood bootstrap values ≥ 75% are shown. The tree is rooted with Pyricularia grisea M83.
-

Figure 2.
Disease symptoms, morphological characters and pathogenicity of Candidacolonium agrostis. (a) Summer patch-like disease on creeping bentgrass putting green. (b) Front and back of colony on PDA plate after 14-d incubation. (c) Hyphopodia. (d), (e) Creeping bentgrass top growth one month after inoculation with sterile and 13g11 inoculum, respectively. (f), (g) Creeping bentgrass roots one month after inoculation with sterile and 13g11 inoculum, respectively.
-

Figure 3.
Total mycelial growth (colony diameter, mm) of Candidacolonium agrostis (13g11 and JH1) and Magnaporthiopsis poae (Y15 and S24) isolates on PDA after 5 d of incubation in the dark at temperatures of 4, 20, 26, 29 and 37 °C. Error bars represent the mean ± standard deviation.
-
Species name Isolate ID ITS LSU SSU MCM7 RPB1 TEF1 Buergenerula spartinae ATCC 22848a JX134666 DQ341492 DQ341471 JX134706 JX134720 JX134692 Candidacolonium agrostisb 13g11 OM910761 OM910782 OM900029 OM938227 OM938229 OM938231 Candidacolonium agrostis 13g12 OM910762 OM910783 OM900030 OM938228 OM938230 OM938232 Candidacolonium agrostis JH1 OP811262 OP811312 OP811273 OP957424 OP957426 OP957428 Candidacolonium agrostis JH2 OP811263 OP811313 OP811274 OP957425 OP957427 OP957429 Candidacolonium cynodontis HP24-3 KJ855497 KM401637 KP129316 KP007344 KP268919 KP282703 Candidacolonium cynodontis HP38-4 KJ855498 KM401638 KP129317 KP007345 KP268920 KP282704 Gaeumannomyces graminis var. graminis GSGC15-4 KJ855495 KM401635 KP129314 KP007342 KP268917 KP282701 Gaeumannomyces graminis var. avenae CBS 187.65 JX134668 JX134680 JX134655 JX134708 JX134722 JX134694 Gaeumannomyces graminis var. tritici M55 JF414850 JF414900 JF414875 JF710395 JF710445 JF710420 Gaeumannomyces paulograminis RS7-1 KJ855507 KM401647 KP129326 KP007350 KP268929 KP282713 Magnaporthiopsis cynodontis RS7-2 KJ855508 KM401648 KP129327 KP007351 KP268930 KP282714 Magnaporthiopsis cynodontis KR10-6 KJ855499 KM401639 KP129318 KP007357 KP268921 KP282705 Magnaporthiopsis cynodontis TPC5-3 KJ855514 KM401654 KP129333 KP007356 KP268936 KP282720 Magnaporthiopsis incrustans WW3-5 KJ855515 KM401655 KP129334 KP007332 KP268937 KP282721 Magnaporthiopsis panicorum CM10S2 KF689644 KF689634 KF689594 KF689604 KF689614 KF689624 Magnaporthiopsis poae TAP41 KJ855512 KM401652 KP129331 KP007354 KP268934 KP282718 Magnaporthiopsis rhizophila M23 JF414834 JF414883 JF414858 JF710384 JF710432 JF710408 Nakataea oryzae M21 JF414838 JF414887 JF414862 JF710382 JF710441 JF710406 Nakataea oryzae M69 JX134672 JX134684 JX134658 JX134712 JX134726 JX134698 Nakataea oryzae M71 JX134673 JX134685 JX134659 JX134713 JX134727 JX134699 Ommidemptus affinis ATCC 200212 JX134674 JX134686 JX134660 JX134714 JX134728 JX134700 Pseudophialophora cynodontis RW3-4 KJ855509 KM401649 KP129328 KP007352 KP268931 KP282715 Pseudophialophora eragrostis CM12M9 KF689648 KF689638 KF689598 KF689608 KF689618 KF689628 Pseudophialophora eragrostis CM20M5-2 KF689647 KF689637 KF689597 KF689607 KF689617 KF689627 Pseudophialophora panicorum CM9S6 KF689651 KF689641 KF689601 KF689611 KF689621 KF689631 Pseudophialophora schizachyii AL2M1 KF689649 KF689639 KF689599 KF689609 KF689619 KF689629 Pyricularia grisea M83 JX134671 JX134683 JX134657 JX134711 JX134725 JX134697 Slopeiomyces cylindrosporus CBS 610.75 JX134667 DQ341494 DQ341473 JX134707 JX134721 JX134693 a ATCC = American Type Culture Collection, Manassas, Virginia; CBS = Centraalbureau voor Schimmelcultures, Utrecht, the Netherlands. b Species names in boldface indicate newly submitted sequences from this study. Table 1.
Species name, isolate IDs, and GenBank accession numbers for fungal isolates used for phylogenetic analyses in this study.
Figures
(3)
Tables
(1)