-
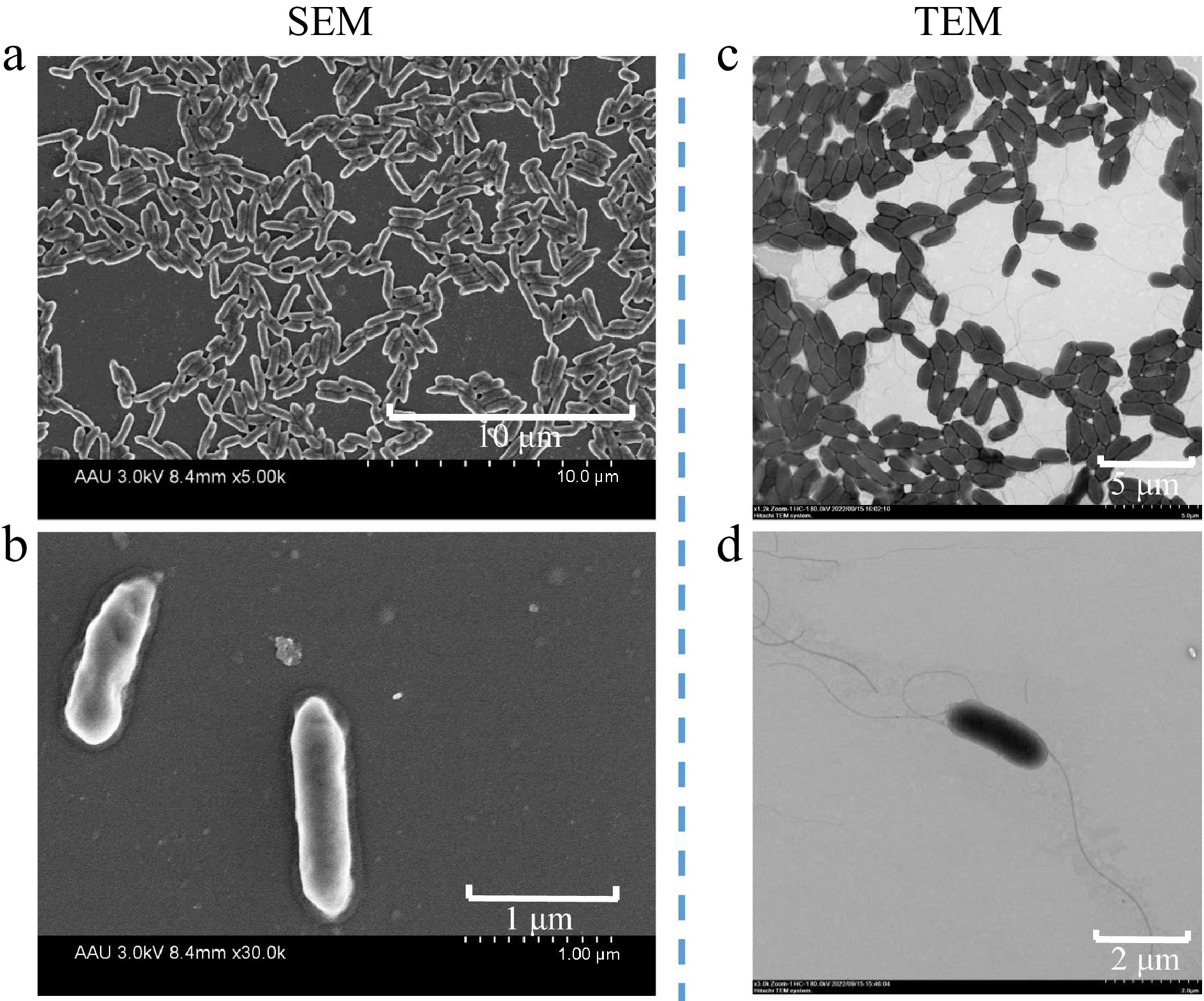
Figure 1.
Scanning electron micrograph (SEM) of strain V4 grown for 10 h in LB solid medium, acceleration voltage of 3.0 kv, (a) 8.4 mm × 5.00 k, scale bar 10 μm, (b) 8.4 mm × 30.0 k, scale bar 1 μm. Transmission electron micrograph (TEM) showing cells of strain V4 grown for 10 h in LB solid medium, with flagella at both ends, acceleration voltage of (c) 80 kv, × 1.2 k, scale bar 5 μm, (d) × 3.0 k, scale bar 2 μm.
-
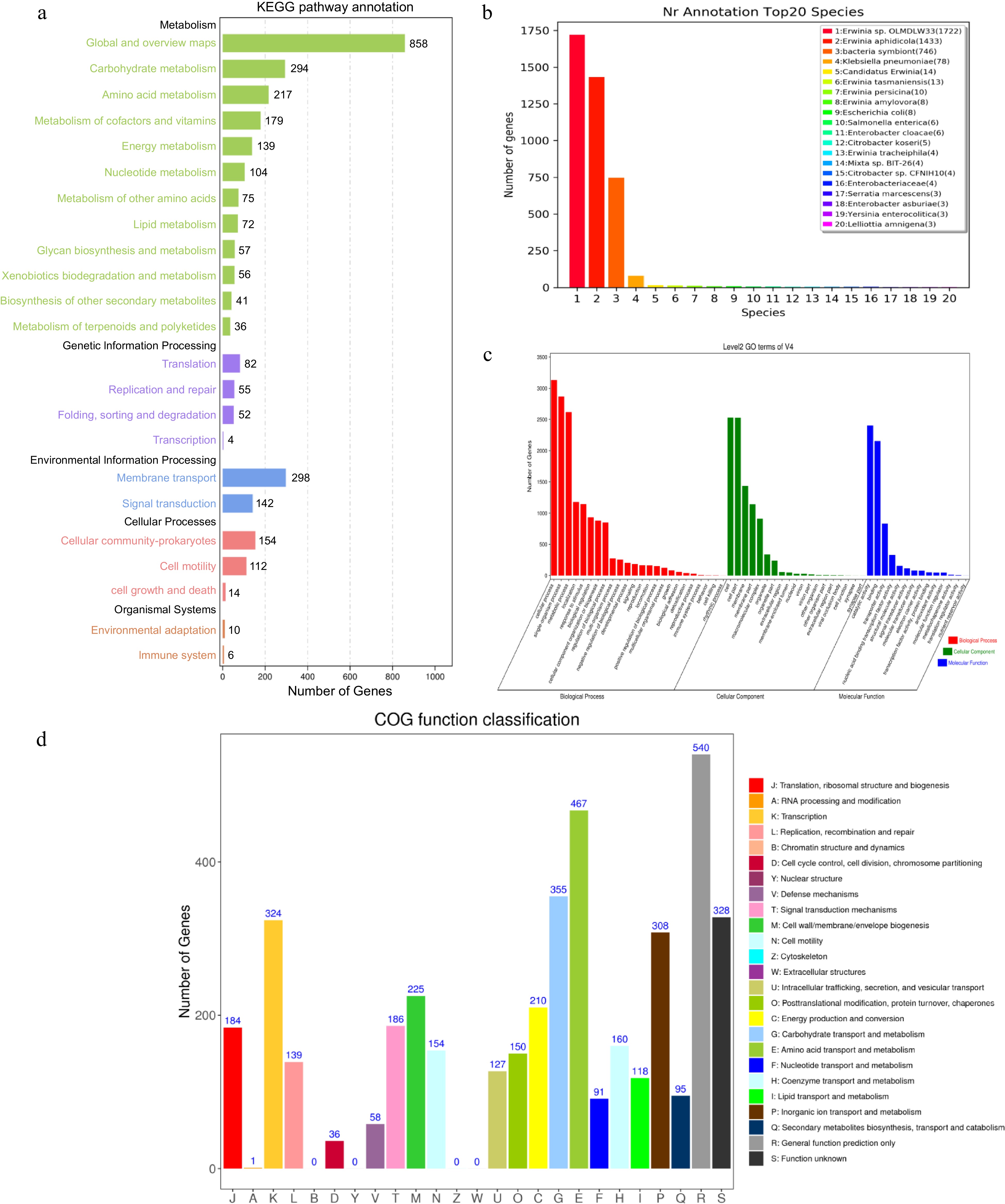
Figure 2.
The V4 chromosome genomic annotation information of gene function based on (a) KEGG, (b) NR, (c) GO and (d) COG databases.
-
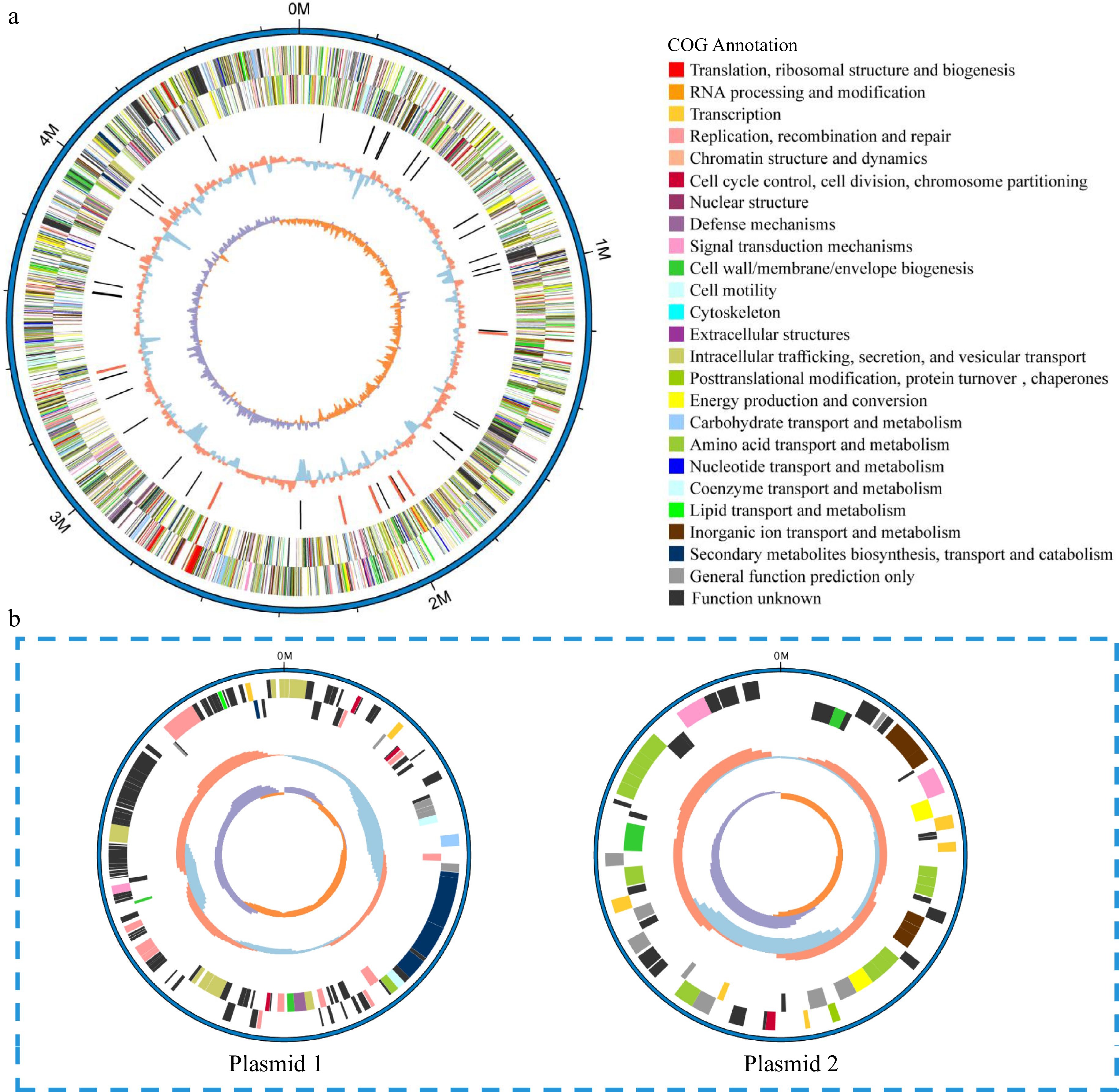
Figure 3.
Circular representation of the (a) chromosome and (b) plasmids of V4 strain. Circles (from outside to inside): forward strand genes and reverse strand genes (annotation information of gene function based on COG databases), ncRNA (black, tRNA, red, rRNA), GC content (red, > mean value of GC content, blue, < mean value of GC content), GC skew (positive and negative values being indicated with purple and orange colors respectively) (purple, > 0, orange, < 0).
-
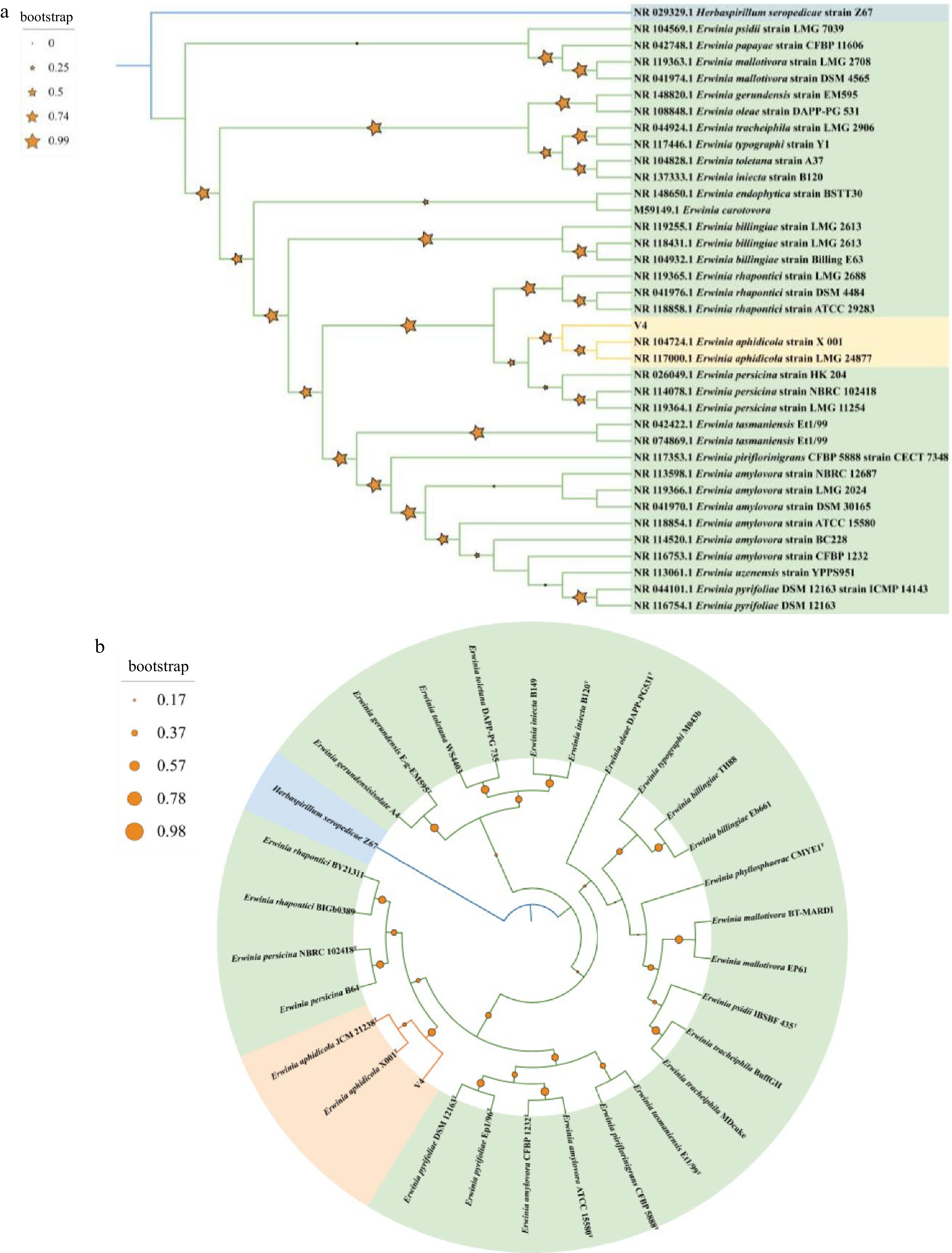
Figure 4.
(a) Maximum-likelihood phylogenomic tree based on 16S rRNA gene sequences among V4 strain and Erwinia genus strains. Herbaspirillum seropedicae Z67T was used as outgroup. Bootstrap values are shown in tree branches calculating for 1000 subsets. (b) Whole-genome-based phylogenomic tree of V4 strain, available Erwinia genus strains and Herbaspirillum seropedicae Z67T. Support values are shown in tree branches.
-
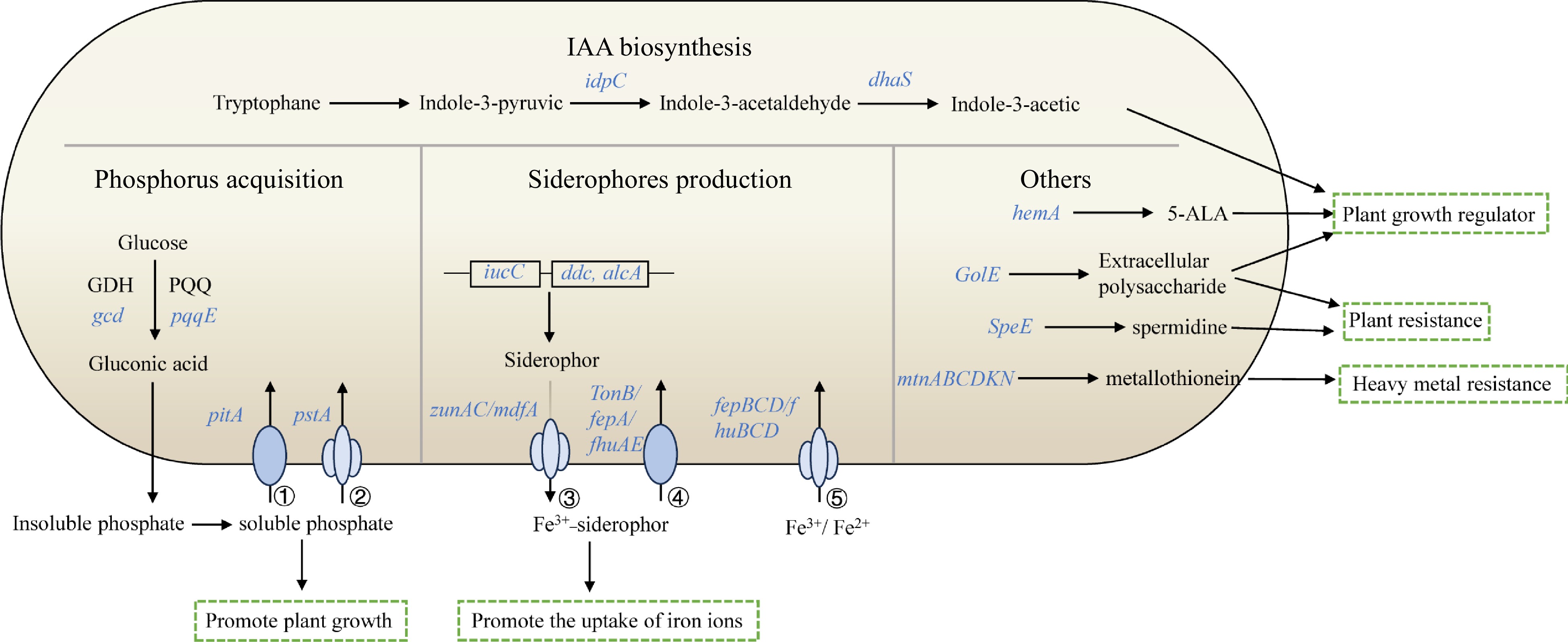
Figure 5.
Schematic overview of main plant growth-promoting traits in V4. The depicted pathways were predicted based on the genomic data of V4. Details are available in Supplemental Table S2. These include IAA biosynthesis, phosphorus acquisition, siderophores production and others. Individual pathways were denoted by single-headed arrows, genes were shown in blue italics. Abbreviations: GDH (glucose dehydrogenase); PQQ (pyrroloquinoline quinine); 5-ALA (5-Aminolevulinic acid); ① low-affinity phosphate transport system, Pit; ② high-affinity phosphate transport system, Pst; ③ siderophore transporter; ④ siderophore outer membrane receptor proteins; ⑤ ABC-type Fe2+/Fe3+-hydroxamate transport protein.
-
Chromosome Plasmid 1 Plasmid 2 Size (bp) 4,697,109 160,141 71,044 G+C content (%) 56.68% 53.46% 54.25% Total protein-coding genes 4189 131 64 Total length (bp) 4,080,951 119,871 55,104 G + C content (%) 57.94% 54.65% 56.67% No. of rRNA operons 22 0 0 No. of tRNAs 84 0 0 No. of sRNAs 31 0 0 Gene islands 4 0 0 Prophage 2 0 0 TNSS II, III, IV IV 0 Table 1.
General characteristics of the strain V4 genome.
Figures
(5)
Tables
(1)