-
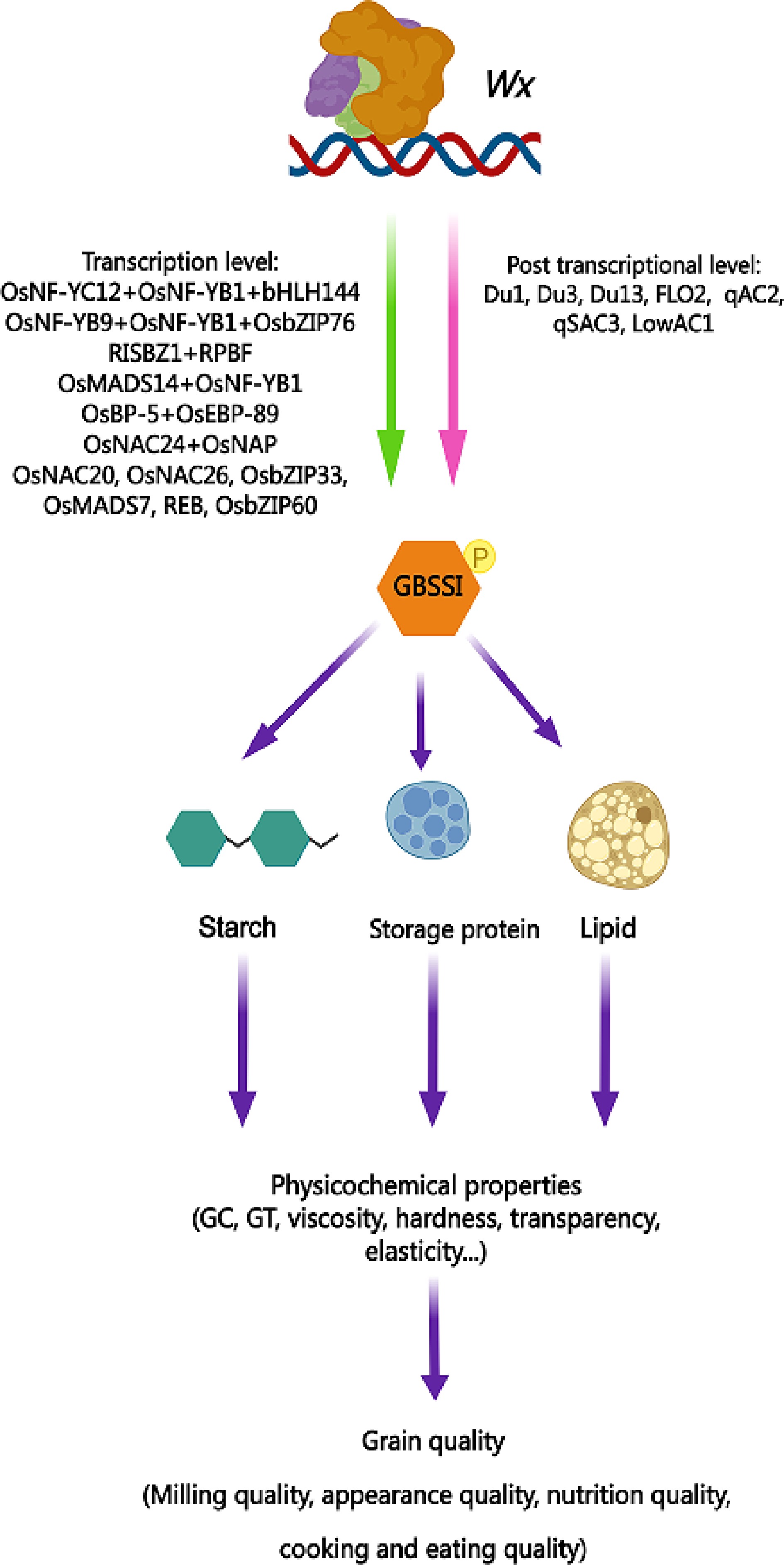
Figure 1.
Pleiotropy of Wx gene in rice quality regulation. This pattern diagram presents the regulatory factors of the Wx gene at both the transcriptional and post-transcriptional levels that have been reported thus far. It also highlights the encoding product GBSSI of the Wx gene, which plays a role in regulating the three major components of rice endosperm (starch, protein, and lipids) and subsequently affects various physicochemical properties of rice grains. The pleiotropic effect of the Wx gene ultimately determines the overall quality of rice from multiple perspectives. GC, gel consistency; GT, gelatinization temperature. Yellow spheres with the letter P indicate phosphorylation.
-
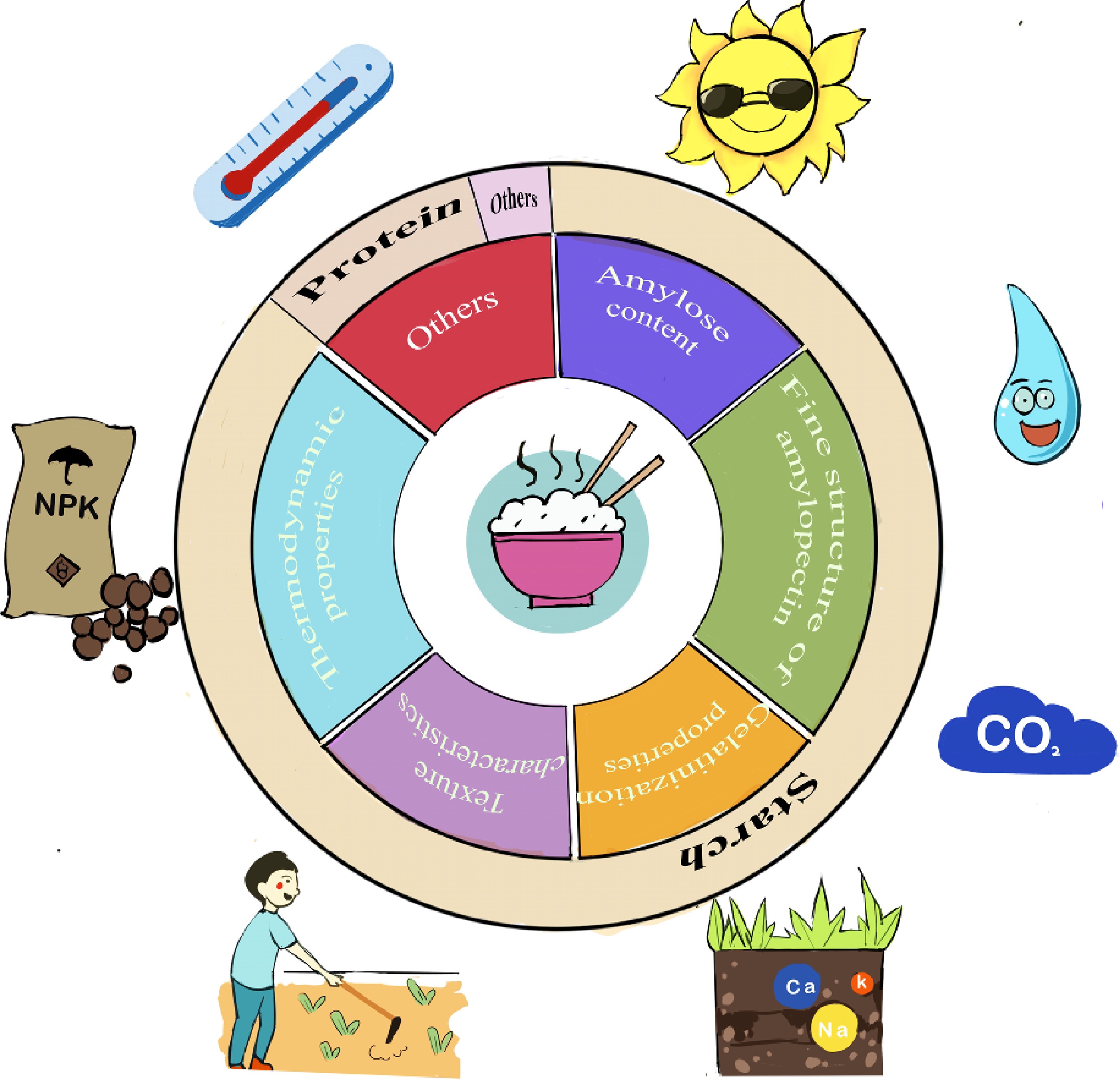
Figure 2.
Determinants of cooking and eating quality of rice and external factors affecting protein content of rice grains. The six parts in the inner circle represent the physical and chemical characteristics or indicators that determine the cooking and eating quality of rice; the three parts in the outer circle represent the material composition that affects the physical and chemical characteristics of rice; the cartoon diagram in the outermost circle represents the ecological factors and environmental factors that affect the grain protein content, from clockwise from 1 o'clock to indicate light, moisture, carbon dioxide, salinity, cultivation, fertilizer and temperature.
-
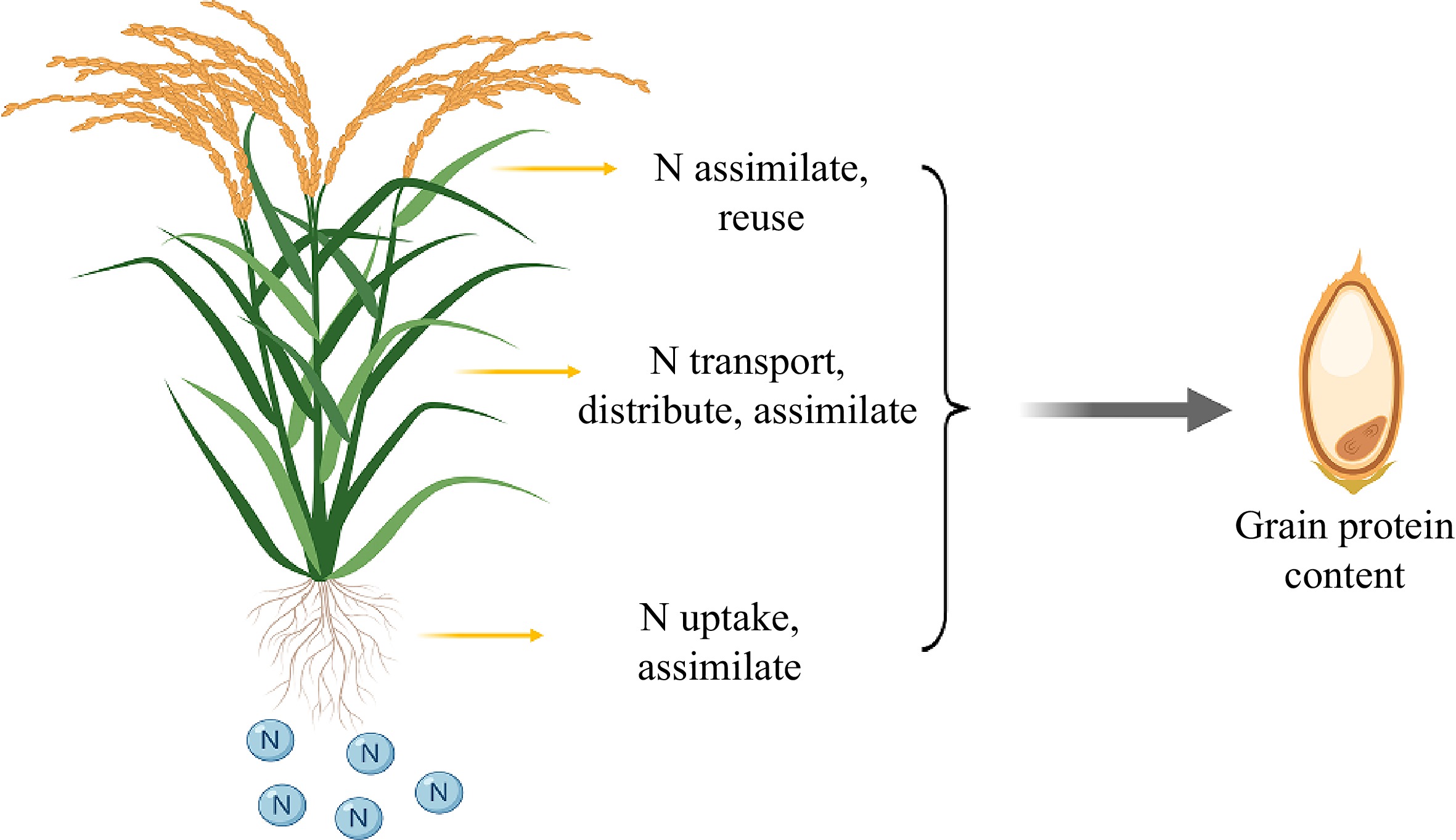
Figure 3.
The nitrogen utilization ability of rice may determine the protein content of grains. Each step of nitrogen utilization in rice, which includes absorption, transportation, assimilation, distribution, and reuse, can potentially impact the final grain protein content.
-
Classification Name Gene ID Effect of gene knockout
or knockdown on grain
protein contentAnnotation Biochemical metabolism flo4/OsPPDKB/OsC4PPDK LOC_Os05g33570 Increased[130] Pyruvate, phosphate dikinase OsSSIIIa/Flo5/SS3a LOC_Os08g09230 Increased[103] Starch synthase III FLO8/OsUgp1 LOC_Os09g38030 Increased[131] UTP--glucose-1-phosphate uridylyltransferase FLO12/OsAlaAT1 LOC_Os10g25130 Increased[132,133] Aminotransferase FLO15/OsGLYI7 LOC_Os05g14194 Increased[134] Glyoxalase family protein FLO16 LOC_Os10g33800 Increased[135] lactate/malate dehydrogenase FSE1 LOC_Os08g01920 Increased[136] Phospholipase-like protein OsAGPL2/OsAPL2/shr1/GIF2/osagpl2-3 LOC_Os01g44220 Decreased[137] ADP-glucose pyrophosphorylase large subunit 2 OsBEIIb/be2b LOC_Os02g32660 Unknown[138] Starch branching enzyme IIb Pho1 LOC_Os03g55090 Unknown[139] Plastidial phosphorylase OsGINT1/FSE6 LOC_Os05g46260 Increased[140] Glycosyltransferase OsPK2/OsPKpα1 LOC_Os07g08340 Increased[141] Plastidic pyruvate kinase GIF1/OsCIN2 LOC_Os04g33740 Unknown[142] Glycosyl hydrolases PDIL1-1 LOC_Os11g09280 Increased[143] Protein disulphide isomerase-like enzyme OsACS6/SSG6 LOC_Os06g03990 Unknown[144] Aminotransferase PFPβ/PFP1 LOC_Os06g13810 Unknown[145] Pyrophosphate-fructose 6-phosphate
1-phosphotransferase subunit betaFLO19 LOC_Os03g48060 Increased[28] Class I glutamine amidotransferase FLO19 LOC_Os04g02900 Decreased[146] Plastid-localized pyruvate dehydrogenase complex E1 component subunit α1 FLO23/OsF2KP2 LOC_Os03g18310 Decreased[147] Fructose-6-phosphate-2-kinase/fructose-2, 6-bisphosphatase OsDPE1 LOC_Os07g43390 Unknown[148] Disproportionating enzyme Transcriptional regulation and protein interaction OsNF-YC12 LOC_Os10g11580 Increased[19,149] CCAAT-box-binding transcription factor OsNF-YB1/OsHAP3K/OsEnS-41 LOC_Os02g49410 Increased[19] Nuclear transcription factor Y subunit B bHLH144 LOC_Os04g35010 Increased[19] Helix-loop-helix DNA-binding domain containing protein RISBZ1/OsbZIP58 LOC_Os07g08420 Decreased[150] bZIP transcription factor REB/OsbZIP33/RISBZ2 LOC_Os03g58250 Unknown[151] bZIP transcription factor RPBF/OsDof3/OsDof-10/OsDof7 LOC_Os02g15350 Decreased[150,152] Dof transcription factor FLO2 LOC_Os04g55230 Unknown[153] Tetratricopeptide repeat domain containing protein FLO6 LOC_Os03g48170 Increased[154] CBM48 domain-containing protein OsGBP LOC_Os02g04330 No change[29] GBSS-binding protein FLO7 LOC_Os10g32680 Unknown[155] DUF1388 domain protein OsHsp70cp-2/cpHSP70-2/ flo11-2/ FLO11 LOC_Os12g14070 Unknown[156] Plastid heat shock protein 70 RSR1 LOC_Os05g03040 Unknown[157] Transcription factor of the AP2/EREBP family OsNAC20; OsNAC26 LOC_Os01g01470; LOC_Os01g29840 Decreased when knock out together[18] NAC transcription factor OsNAC23/ONAC023 LOC_Os02g12310 Decreased when gene knock out and increased when gene overexpression[158] NAC transcription factor OsNAC24/OsNAC024 LOC_Os05g34310 Unknown[17] NAC transcription factor OsNAC127 LOC_Os11g31340 Unknown[159] NAC transcription factor OsNAC129 LOC_Os11g31380 Unknown[159,160] NAC transcription factor Du13/TL1 LOC_Os06g48530 No change[12] C2H2 zinc finger protein OsbZIP60/O3/OPAQUE3 LOC_Os07g44950 Decreased[24] Basic leucine zipper transcription factor OsMADS6/MFO1 LOC_Os02g45770 Increased[161] MADS-box transcription factor OsMADS29 LOC_Os02g07430 Unknown[162] MADS-box transcription factor OsMADS14 LOC_Os03g54160 Unknown[21] MADS-box transcription factor Epigenetics OsROS1/ROS1a/DNG702 LOC_Os01g11900 Increased[163] DNA demethylase OsCADT1/FLO20/SHMT4 LOC_Os01g65410 Decreased[164] Serine hydroxymethyltransferase Energy supply FLO10 LOC_Os03g07220 Increased[165] Pentatricopeptide repeat protein FLO14/OsNPPR3 LOC_Os03g51840 Unknown[166] Pentatricopeptide repeat protein FLO18 LOC_Os07g48850 Decreased[167] Pentatricopeptide repeat protein OGR1 LOC_Os12g17080 Unknown[168] Pentatricopeptide repeat–DYW protein FGR1/OsNPPR1 LOC_Os08g19310 Unknown[169] Pentatricopeptide repeat protein FLO13/OsNDUFA9 LOC_Os02g57180 Unknown[169] Mitochondrial complex I subunit FLO22 LOC_Os07g08180 Unknown[170] P-type pentatricopeptide repeat (PPR) protein Material transport ESG1 LOC_Os04g46700 Decreased[171] Bcterial-type ABC (ATP-binding cassette) lipid transporter OsBT1 LOC_Os02g10800 Unknown[172] ADP-Glucose Transporter OsBip1/BiP3 LOC_Os02g02410 Decreased[173] Endoplasmic riculum caperone OsLTPL36 LOC_Os03g25350 Decreased[174] Lipid transfer protein OsRab5a/gpa1/glup4 LOC_Os12g43550 No change, but pro-glutelin accumulation[175] Small GTPase Sar1a; Sar1b; Sar1c LOC_Os01g23620; LOC_Os12g37360; LOC_Os01g15010 Unknown but pro-glutelin accumulation when three genes knockdown together[176] Small GTPase GPA3 LOC_Os03g61950 Increased and pro-glutelin accumulation[177] Regulator of post-Golgi vesicular Traffic GPA4/GLUP2/GOT1B LOC_Os03g11100 Decreased but pro-glutelin accumulation[178] Golgi Transport 1 GPA5 LOC_Os06g43560 Unknown but pro-glutelin accumulation[179] Rab5a Effector OsVPS9A/GPA2/GLUP6/GEF LOC_Os03g15650 Decreased but pro-glutelin accumulation[180] Guanine nucleotide exchange factor GPA6/OsNHX5 LOC_Os09g11450 No change, but pro-glutelin accumulation[181] Vacuolar Na+/H+ antiporter Function unknown SSG4 LOC_Os01g08420 Unknown[182] Unknown function protein Table 1.
Reported floury endosperm genes that may affect grain protein content.
-
Name Gene ID Effect of altered gene function on grain protein content Annotation MYB61/qNLA1/qCel1 LOC_Os01g18240 Unknown[183] MYB family transcription factor OsGRF4/GS2/GL2/PT2/LGS1/GLW2 LOC_Os02g47280 Increased when gene overexpression[108] Growth-regulating factor TOND1 LOC_Os12g43440 Unknown[184] Unkonwn function protein OsDEP1/DN1/qPE9-1/qNGR9 LOC_Os09g26999 No change when gene overexpression[185] Gγ subunit OsTCP19 LOC_Os06g12230 Unknown[186] Class-I TCP transcription factor SMOS1/shb/RLA1/NGR5 LOC_Os05g32270 Unknown[187] GRAS protein OsNPF6.1 LOC_Os01g01360 Unknown[188] Nitrate transporter OsNAC42 LOC_Os09g32040 Unknown[188] No apical meristem protein OsNR2/qCR2 LOC_Os02g53130 Unknown[189] Nitrate reductase OsNRT1.1B/OsNPF6.5/qCHR-10 LOC_Os10g40600 Unknown[190] Peptide transporter PTR2 Ghd7/E1/Hd4 LOC_Os07g15770 Unknown[191] CCT motif family protein ARE1 LOC_Os08g12780 Unknown[192] Chloroplast envelope membrane protein OsCCA1/OsLHY/Nhd1 LOC_Os08g06110 Decreased when gene expression enhanced[107] MYB transcription factor Ef-cd Unknown[193] Long noncoding RNA OsAtg8/OsATG8a LOC_Os07g32800 Increased when gene overexpression[194] Autophagy-related protein OsPIN9 LOC_Os01g58860 Unknown[195] Auxin efflux transporter OsNLP4 LOC_Os09g37710 Unknown[196,197] NIN-like protein OsNLP3 LOC_Os01g13540 Unknown[198] NIN-like protein OsNLP1 LOC_Os03g03900 Unknown[199] NIN-like protein OsAAP6/qPC1 LOC_Os01g65670 Increased when gene overexpression[25] Amino acid permease OsAAP10 LOC_Os02g49060 Decreased when gene knock out[200] Amino acid permease OsAAP5 LOC_Os01g65660 Unknown[201] Amino acid permease OsAAP3 LOC_Os06g36180 Increased when gene overexpression[202] Amino acid permease OsAAP1 LOC_Os07g04180 Increased when gene overexpression and decreased when gene interference[203] Amino acid permease OsAMT1;1/OsAMT1-1 LOC_Os04g43070 Unknown[204] High-affinity ammonium transporter OsAMT2;1 LOC_Os05g39240 Unknown[205] Ammonium transporter OsGS1/OsGS1;1/OsGLN1;1/λGS28 LOC_Os02g50240 Decreased when OsGS1;1b overexpression[206] Glutamine synthetase OsGS2/OsGLN2/λGS31 LOC_Os04g56400 Increased when concurrent overexpression of OsGS1 and OsGS2[207] Glutamine synthetase OsAMT1;3/OsAMT1.3/OsAMT1;2 LOC_Os02g40710 Increased when gene overexpression[208] Ammonium transporter OsGOGAT1 LOC_Os01g48960 Increased when gene overexpression[208] Glutamate synthetase 1 OsAS1/ OsASN1 LOC_Os03g18130 Increased when gene overexpression[209] Asparagine synthetase OsSHM1/OsSHMT1 LOC_Os03g52840 Unknown[210] Serine hydroxymethyltransferase 1 OsENOD93-1 LOC_Os06g05010 Unknown[211] Early nodulin 93 ENOD93 protein OsNRT2.3/OsNRT2.3a/OsNRT2.3b LOC_Os01g50820 Unknown[212] High-affinity nitrate transporter OsNAR2.1 LOC_Os02g38230 Unknown[213] Partner protein for high-affinity nitrate transport OsNRT2.1 LOC_Os02g02170 Unknown[214] High-affinity nitrate transporter OsNRT1.1A/OsNPF6.3 LOC_Os08g05910 Unknown[215] Nitrate transporter DNR1 LOC_Os01g08270 Decreased when gene knock out[216] Amino transferase qSBM1 LOC_Os01g65120 Unknown[217] Peptide transporter OsRBCS2 LOC_Os12g17600 Unknown[218] Small subunit of Rubisco OSA1 LOC_Os03g48310 Unknown[219] Plasma membrane H+-ATPase OsDREB1C LOC_Os06g03670 Increased when gene overexpression[111] AP2/EREBP transcription factor OsPTR6/OsNPF7.3 LOC_Os04g50950 Unknown[220] Peptide transporter OsMADS1/LHS1/AFO/LGY3/GW3p6 LOC_Os03g11614 Unknown[221,222] MADS-domain transcription factor Table 2.
Reported nitrogen use efficiency genes that may also affect grain protein content.
Figures
(3)
Tables
(2)