-
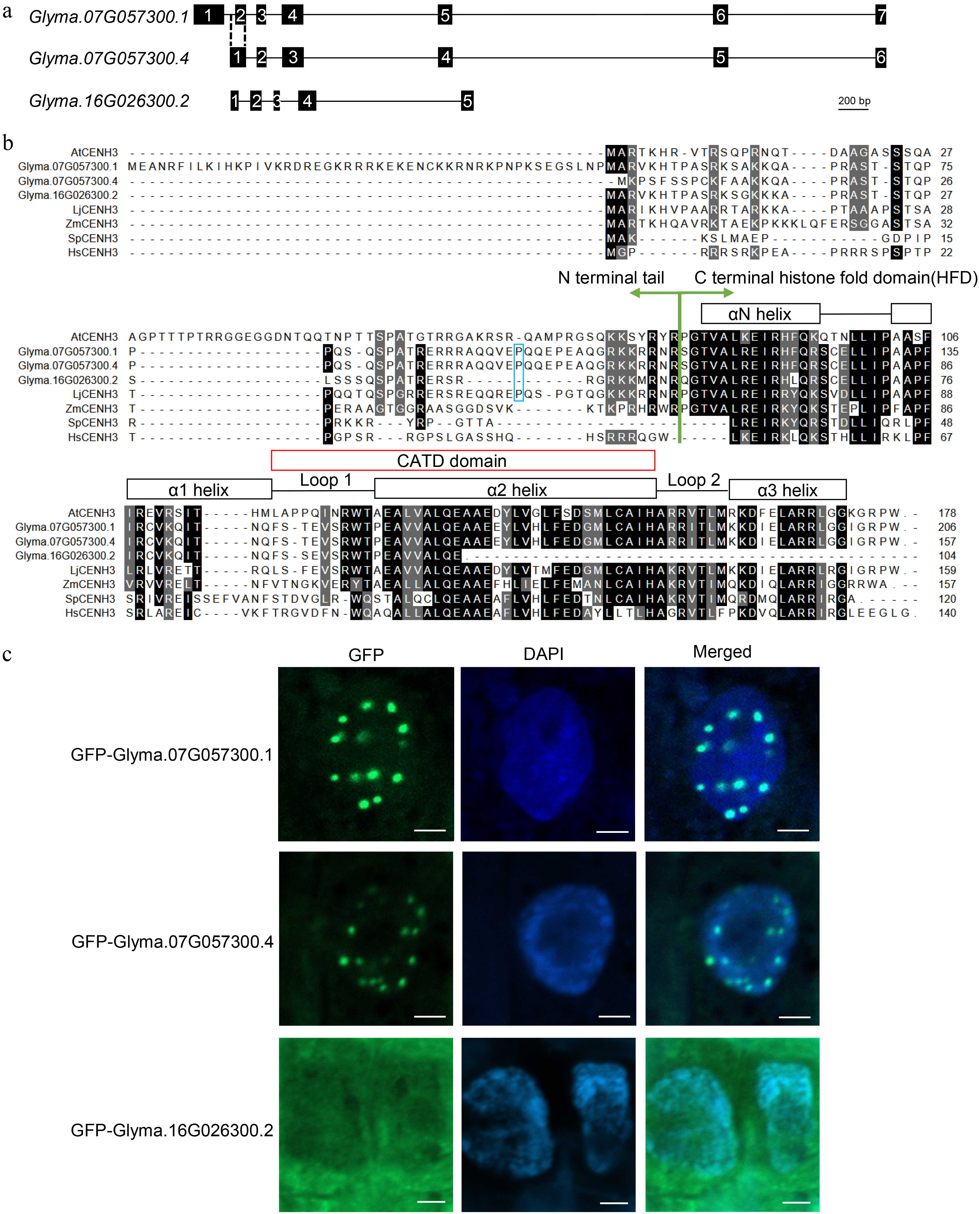
Figure 1.
Identification of CENH3 in soybean. (a) Transcript scheme of Glyma.07G057300 and Glyma.16G026300. (b) Protein alignment of AtCENH3 (Arabidopsis thaliana, NP_001030927.1), Glyma.07G057300.1, Glyma.07G057300.4, Glyma.16G026300.2, LjCENH3 (Lotus japonicus, AHH01567.1), ZmCENH3 (Zea mays, NP_001105520.1), SpCENH3 (Schizosaccharomyces pombe, NP_596473.1) and HsCENH3 (Homo sapiens, NP_001800.1). (c) Subcellular locations of the GFP fusion proteins of Glyma.07G057300.1, Glyma.07G057300.4 and Glyma.16G026300.2. Scale bar = 2 μm.
-
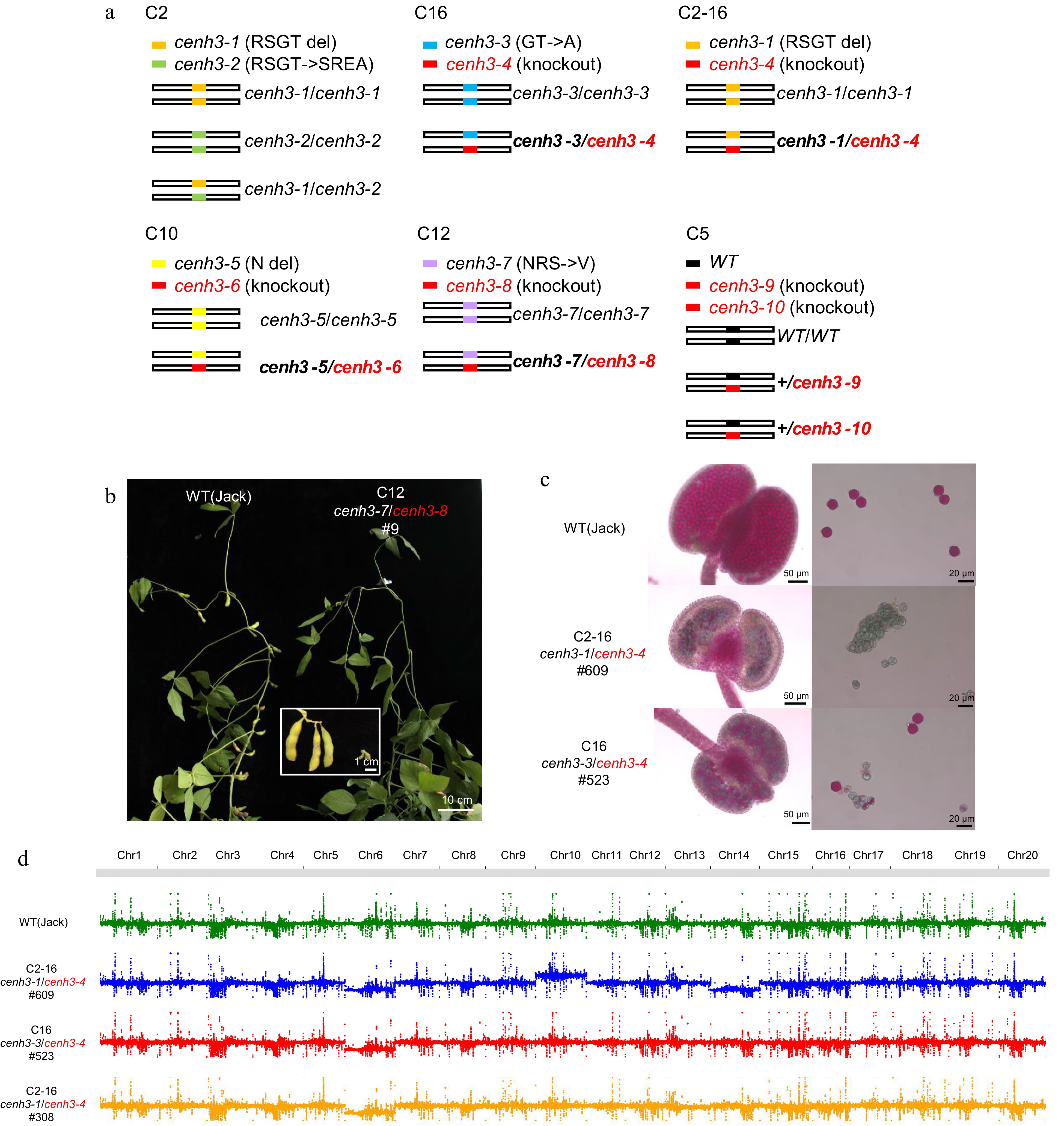
Figure 2.
Heterozygosity of GmCENH3 causes genome instability. (a) Diagram of genotypes of six lines with edited GmCENH3. Null alleles were marked by red font and the null heterozygotes were marked by bold font. (b) A plant with the genotype cenh3-7/cenh3-8 among the progeny of null heterozygote C12 was totally sterile. Scale bar = 10 cm. (c) The inviable pollen in the two sterile plants (#609 and #523) among the progeny of null heterozygotes C2-16 and C16. Pollen of plant #609 with genotype cenh3-1/cenh3-4 were totally inviable, and pollen of plant #523 with genotype cenh3-3/cenh3-4 were partially inviable. Pollen were stained by Alexander staining. (d) #609, #523 and #308 had abnormal chromosome numbers.
-
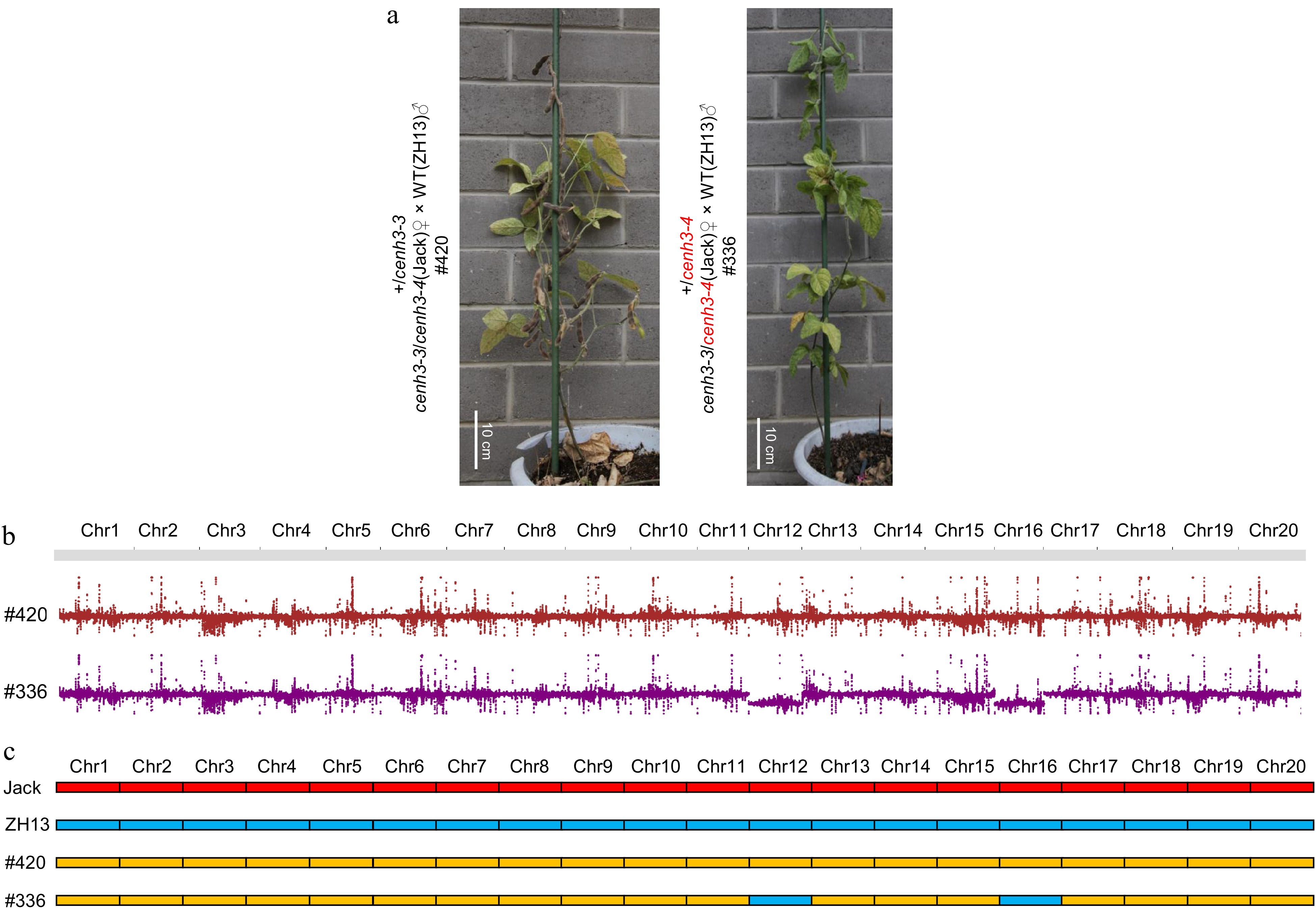
Figure 3.
Female gamete of null allele causes chromosome elimination. (a) 5-month-old F1 plants from the parental null heterozygotes cenh3-3/cenh3-4 and ZH13. #420 was fertile while #336 was sterile. Scale bar = 10 cm. (b) #420 had 40 chromosomes while #336 lost one chromosome 12 and one chromosome 16. (c) The two lost chromosomes in #336 originated from the null heterozygote cenh3-3/cenh3-4. Homozygous molecular markers from Jack were marked as red, and homozygous molecular markers from ZH13 were marked as blue, and heterozygous molecular markers between Jack and ZH13 were marked as yellow.
-
Genotype Number
of plantsExpected number Chi-square test C2 (cenh3-1/cenh3-2) cenh3-1/cenh3-1 17 15 cenh3-2/cenh3-2 15 15 0.9086 cenh3-1/cenh3-2 29 31 C16 (cenh3-3/ cenh3-4 )cenh3-3/ cenh3-4 102 133 0.0016** cenh3-3/cenh3-3 97 66 C2-16 (cenh3-1/ cenh3-4 )F2cenh3-1/ cenh3-4 21 123 <0.0001**** cenh3-1/cenh3-1 163 61 C2-16 (cenh3-1/ cenh3-4 )F3, F4cenh3-1/ cenh3-4 130 174 <0.0001**** cenh3-1/cenh3-1 131 87 C10 (cenh3-5/ cenh3-6 )cenh3-5/ cenh3-6 40 46 0.2919 cenh3-5/cenh3-5 29 23 C12 (cenh3-7/ cenh3-8 )cenh3-7/ cenh3-8 102 107 0.5570 cenh3-7/cenh3-7 58 53 C5-4 (+/ cenh3-9 )+/ cenh3-9 45 50 0.3969 +/+ 30 25 C5-40 (+/ cenh3-10 )+/ cenh3-10 55 63 0.2316 +/+ 40 32 Red: null alleles. Table 1.
Progeny segregation of edited-GmCENH3.
-
Crosses Number of F1 plants Number of +/cenh3-x (x = 1,3) Number of +/ cenh3-4 cenh3-1/ cenh3-4 (Jack) ♀ × WT(Wm82) ♂94 94 0 cenh3-1/ cenh3-4 (Jack) ♀ × WT(Jack )♂16 16 0 WT(Jack) ♀× cenh3-1/ cenh3-4 (Jack) ♂28 13 15 cenh3-3/ cenh3-4 (Jack) ♀ × WT(Wm82) ♂70 70 (11 aborted seeds) 0 cenh3-3/ cenh3-4 (Jack) ♀ × WT(ZH13) ♂18 17 (1 aborted seed) 1 Red: null alleles. Table 2.
Transmission efficiencies of cenh3-1, cenh3-3 and cenh3-4.
Figures
(3)
Tables
(2)