-
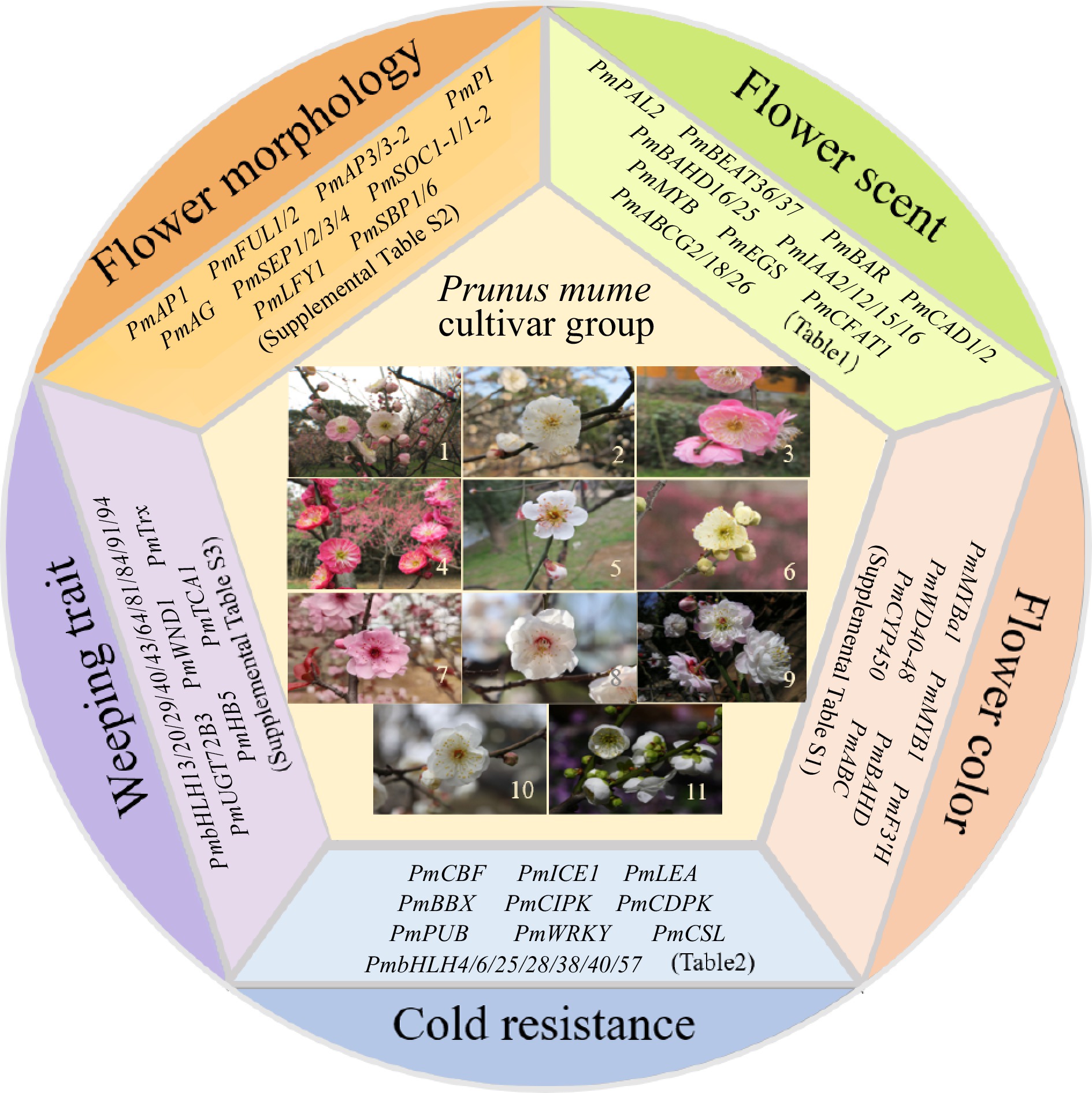
Figure 1.
Eleven cultivar groups of Mei and representative functional genes in five aspects. 1, Versicolor Group; 2, Dragon Group; 3, Pendent Group; 4, Cinnabar Purple Group; 5, Single Flowered Group; 6, Flavescens Group; 7, Blireiana Group; 8, Apricot Mei Group; 9, Pink Double Group; 10, Alboplena Group; 11, Green Calyx Group.
-
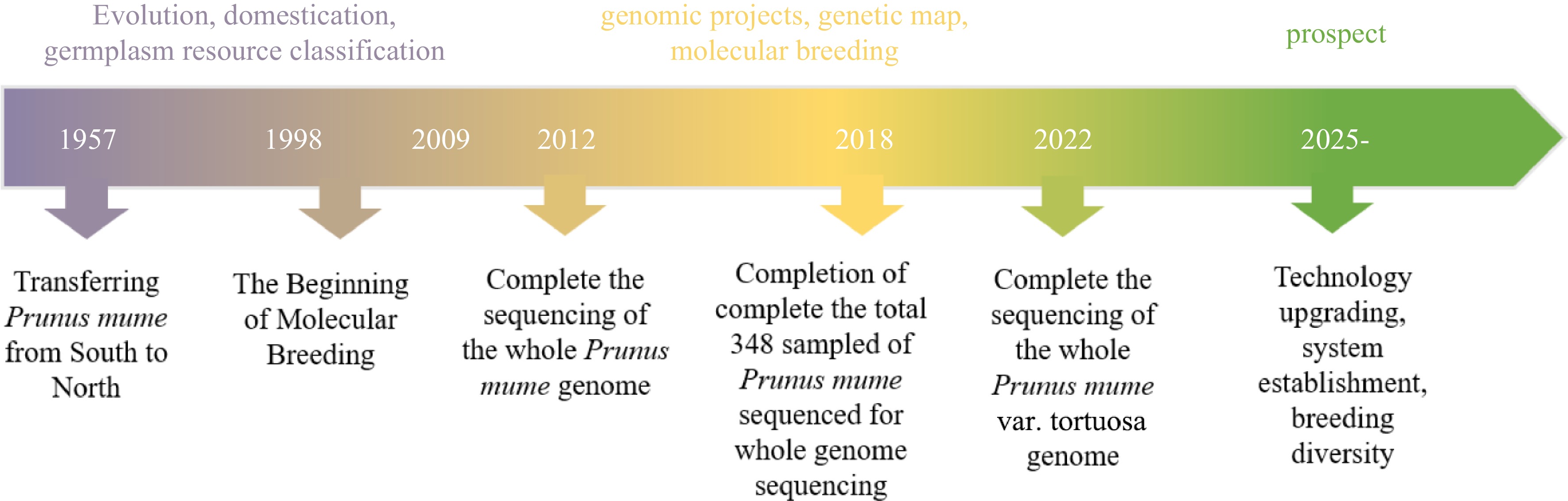
Figure 2.
Timeline of research on P. mume genomics and prospects.
-
Flower scent Gene ID Function description Validation methods Reference PmPAL2 Pm030127 Involved in phenylpropanoids/
benzenoids biosynthesisHS-SPME-GC-MS Methods [32] PmBEAT36/37 Pm011009/Pm011010 Performs a key part in the biosynthesis
of benzyl acetateBioinformatics analysis, expression pattern analysis, plasmid construction, subcellular localization, enzyme activity analysis, GC-MS analysis [11] PmBAR Pm012335/Pm013777/
Pm013782Performs a key part in the biosynthesis
of benzyl acetate, as the key genes responsible for BAR activityIntegrative metabolite, enzyme activity, and transcriptome analysis, plasmid construction, qPCR validation [42] PmBAHD16/25 Pm010996/Pm011009 Plays an important role in promoting
the production of benzyl acetateGC-MS analysis, bioinformatics analysis, expression pattern and WGCNA analysis, validation of transgenic Arabidopsis plants [41] PmIAA2/12/15/16 Pm003529/Pm013416/
Pm013597/Pm020225Involved in the synthesis of benzyl
acetate and cinnamyl acetateBioinformatics analysis, expression pattern analysis [45] PmMYB Pm015692/Pm021211/
Pm025253Engaged in floral fragrance metabolic control via influencing the expression
of downstream genesGC-MS analysis, bioinformatics analysis, expression pattern analysis, subcellular localization, vector construction [18] PmEGS Pm012360 Involved in eugenol biosynthesis Bioinformatics analysis, expression pattern analysis, subcellular localization [43] PmCFAT1 Pm030674 Involved in floral scent metabolism GC-MS analysis, bioinformatics analysis, expression pattern analysis, subcellular localization [34] PmABCG2/18/26 Pm001070/Pm022014/
Pm029602Positively linked with benzaldehyde and benzyl alcohol volatilization rates Bioinformatics analysis, expression pattern analysis, GC-MS analysis of volatile components [44] PmABCG9/13/23 Pm011453/Pm012323/
Pm026080Positively linked with benzaldehyde and benzyl alcohol volatilization rates Bioinformatics analysis, expression pattern analysis, GC-MS analysis of volatile components [44] PmCAD1/2 Pm021215/Pm021214 Play roles in cinnamyl alcohol synthesis GC-MS analysis, bioinformatics analysis, expression pattern analysis, real-time fluorescence quantitative PCR, vector construction [46] Table 1.
Functional validation information of flower scent.
-
Cold resistance Gene ID Function description Validation Methods Reference PmCBF1/2/3/4/5/6 Pm023769/Pm023772/
Pm023773/Pm023775/
Pm023777/Pm027913/Key TF that responses to cold signal qRT-PCR, gene cloning and Yeast 2 Hybrid assays, BiFC assays, promoter cloning and Yeast 1 Hybrid assays [82] PmICE1 − PmCBF express levels are increased in response to a low temperature signal Bioinformatics analysis, expression pattern analysis, subcellular localization, vector construction, Arabidopsis transformation, and low-temperature stress experiments [92] PmLEA10/29 Pm026684/Pm006114 Besponse to cold stress Gene expression analysis, Tobacco transformation and stress tolerance analysis, Relative Water Content (RWC), protein assay and analysis, statistical approach for MDA and REL [86] PmLEA19/20 Pm020945/Pm021811 Response to cold stress; participate in ABA-dependent pathway Gene expression analysis, Tobacco transformation and stress tolerance analysis, Relative Water Content (RWC), protein assay and analysis, statistical approach for MDA and REL [86] PmRS Pm027594/Pm025896 Diminish the negative effects of cold Gene cloning, expression pattern analysis, subcellular localization, transformation of Arabidopsis thaliana, cold resistance analysis, transformation of Mei [107] PmBBX32 Pm013051 Diminish the negative effects of cold Expression pattern analysis, transformed Arabidopsis thaliana, low-temperature stress treatment, physiological index determination [95] PmCIPK5/6/13 Pm001690/Pm018300/
Pm008498Modulating the stress response to cold Bioinformatics analysis, expression pattern analysis, qRT-PCR [103] PmNAC11/20/23/40/
42/48/57/59/60/61/
66/82/85/86/107Pm001403/Pm005783/ Pm006470/Pm011234/
Pm011603/Pm012630/
Pm015876/Pm017550/
Pm018292/Pm018442/
Pm019659/Pm024558/
Pm025184/Pm025307/
Pm025184/Pm025307/
Pm028721Involved in the cold-stress response Bioinformatics analysis, expression pattern analysis, qRT-PCR [100] PmbHLH4/6/25/28/ 38/40/57 Pm002111/Pm002283/
Pm008898/Pm016406/ Pm018355/Pm023237Play a critical part in the resistance to low temperature stress Bioinformatics analysis, expression pattern analysis, qRT-PCR [93,94] PmPUB1/3 Pm006753/Pm009248 Play a significant part in the regulatory network connected to low temperature stress Bioinformatics analysis, expression pattern analysis, qRT-PCR, overexpression of tobacco, low-temperature treatment, physiological index measurement [87] PmWRKY18 Pm005698 Play a significant part in the regulatory network connected to low temperature stress, sensitive to ABA treatment Bioinformatics analysis, expression pattern analysis, qRT-PCR, overexpression of tobacco, low-temperature treatment, physiological index measurement [87] PmWRKY57 LOC103321497 Function in improving cold tolerance of plants Cloning and sequence analysis, subcellular localization, transformation of A. thaliana, determination of plant physiological index, expression analysis of genes [102] PmSOD3 Pm003436 Had important regulatory roles in cold acclimation process Physiological index determination, section observation, tissue browning, ion leakage rate, infrared thermal imaging technology, and freeze thaw detection sensors [109] PmPOD2/19 Pm000967/Pm022119 Had important regulatory roles in cold acclimation process Physiological index determination, section observation, tissue browning, ion leakage rate, infrared thermal imaging technology, and freeze thaw detection sensors [109] PmNCED3/8/9 Pm005153/Pm011164/
Pm016267Significant role in the plant's response to cold stress Bioinformatics analysis, expression pattern analysis, qRT-PCR [108] PmCSL − Significantly improved the freezing tolerance of transgenic plants Bioinformatics analysis, expression pattern analysis, ATAC sequencing, gene cloning and gene expression analysis, plant transformation and low temperature treatment [97] PmHDAC1/6/14 Pm020717/Pm024325/
Pm012683Significantly respond to cold stress Bioinformatics analysis, expression pattern analysis, qRT-PCR [98] PmbZIP 12/31/36/41/48 Pm005288/Pm020080/
Pm021804/Pm025001/
Pm029028Responses to low-temperature stress Bioinformatics analysis, expression pattern analysis, qRT-PCR [99] PmSWEET1/12/13/14 Pm007697/Pm022696/
Pm024167/Pm024554Responses to cold stress Bioinformatics analysis, expression pattern analysis, qRT-PCR [101] PmCDPK14 Pm026757 Play an essential role in resisting low temperature stress Bioinformatics analysis, expression pattern analysis, qRT-PCR, compare between two genomes of Mei [105] PmMAPK3/5/6/20 Pm000966/Pm023935/
Pm027774/Pm014593Significantly respond to cold stress Bioinformatics analysis, expression pattern analysis, qRT-PCR, compare between two genome databases of Mei [104] PmMAPKK2/3/6 Pm027015/Pm015648/
Pm027289Significantly respond to cold stress Bioinformatics analysis, expression pattern analysis, qRT-PCR, compare between two genome databases of Mei [104] PmRCI2s Pm027750/Pm003262/
Pm003263Significantly induced by low temperature Bioinformatics analysis, expression pattern analysis, qRT-PCR [106] Table 2.
Functional validation information of cold resistance.
Figures
(2)
Tables
(2)