-
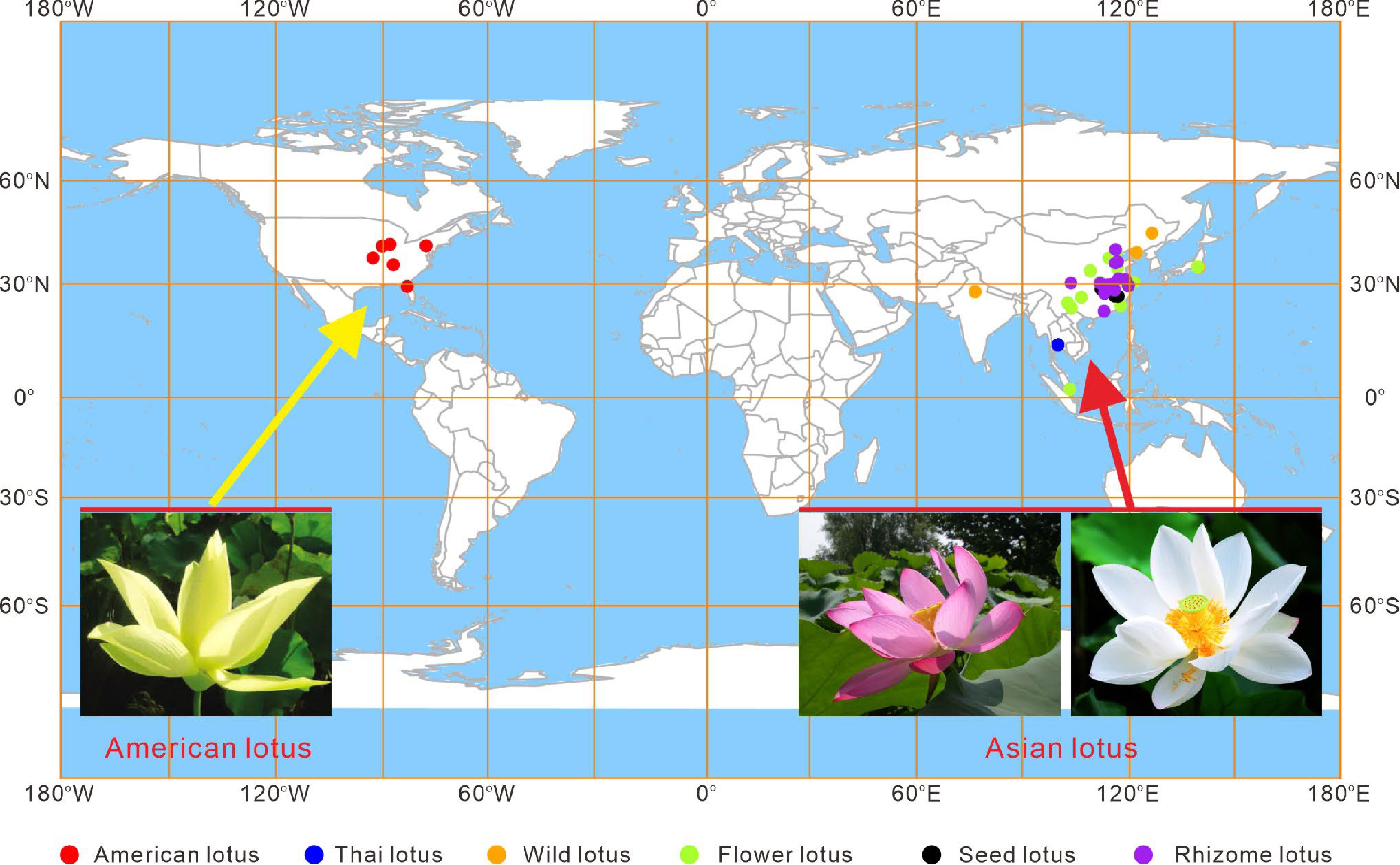
Figure 1.
Geographical distribution of lotus accessions collected in the Wuhan National Lotus Germplasm Bank at the Wuhan Botanical Garden of the Chinese Academy of Sciences. The different colored dots indicate cultivars belonging to different categories.
-

Figure 2.
1-Benzylisoquinoline alkaloids reported in Nelumbo nucifera.
-

Figure 3.
Aporphine-type alkaloids isolated from Nelumbo nucifera.
-

Figure 4.
Bis- and tri- benzylisoquinoline alkaloids isolated from Nelumbo nucifera.
-
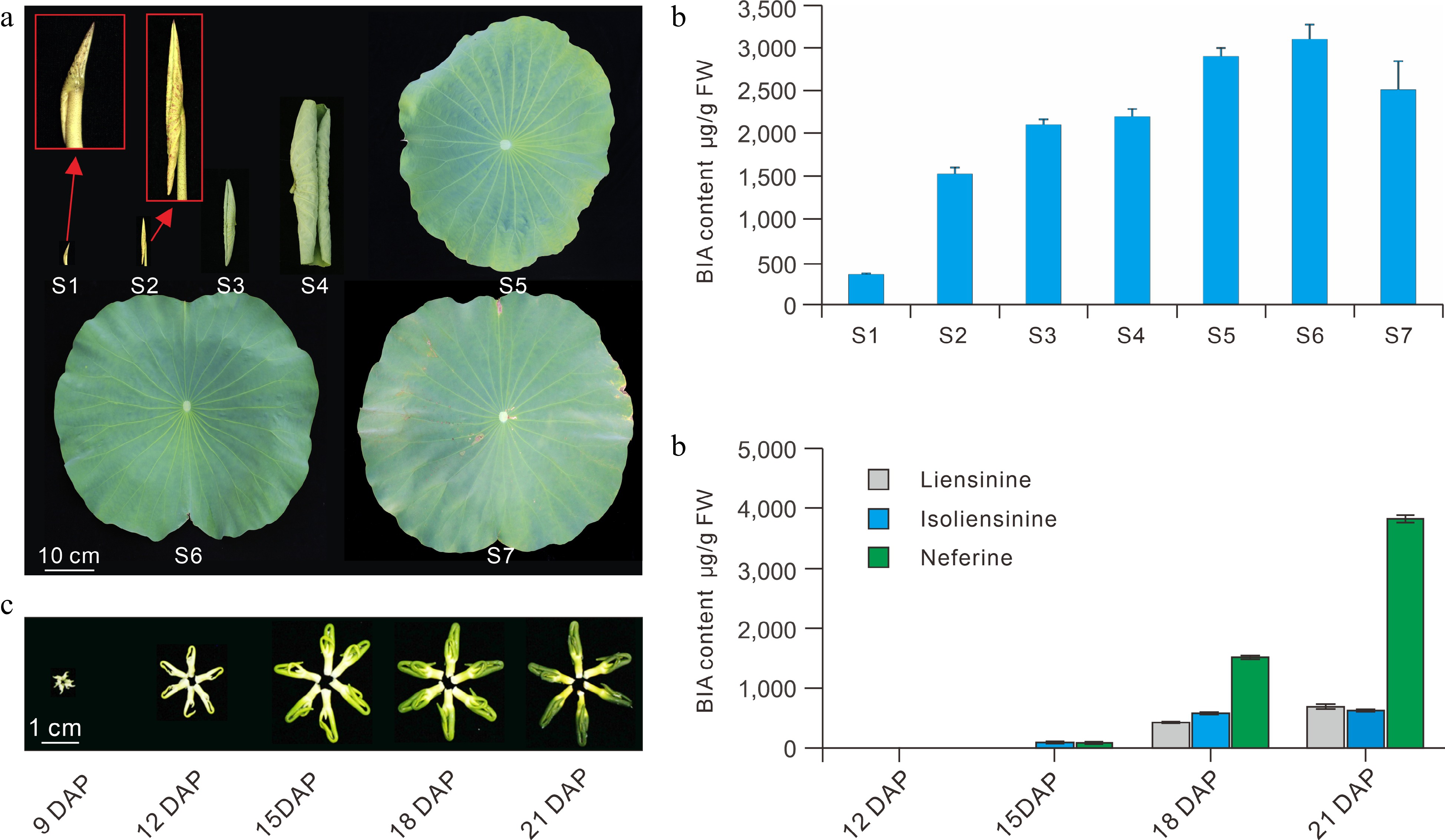
Figure 5.
BIA profiles in the leaf and plumule during development. (a) Lotus leaves showing the seven defined developmental stages. (b) BIA profiles in the seven leaf developmental stages. (c) Lotus plumules showing different developmental stages. (d) BIA content in the plumule at different developmental stages. S, leaf developmental stages; DAP, days post pollination. The figure is modified from previous reports[11,24].
-
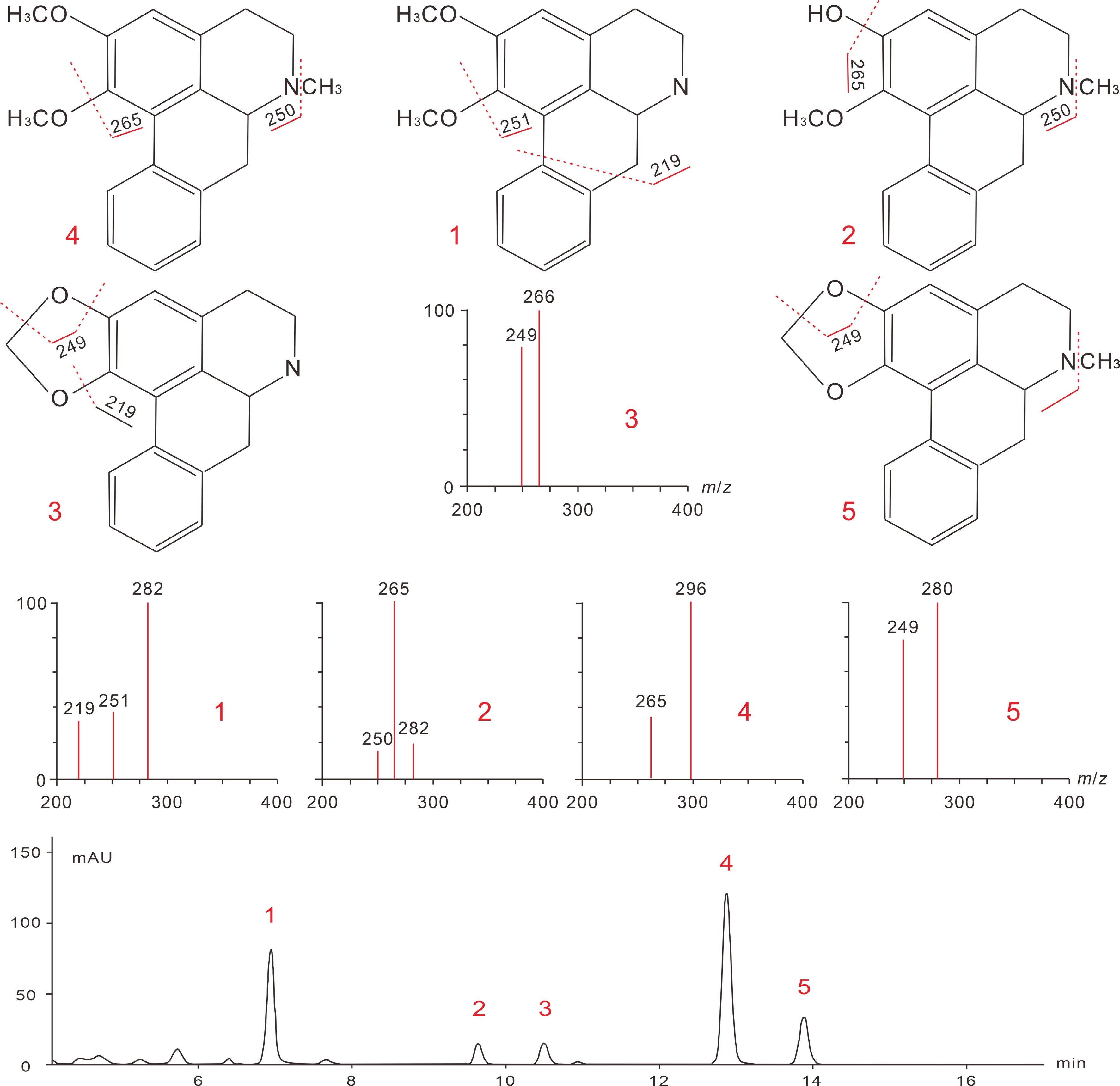
Figure 6.
Analytical mass spectra of lotus leaf BIAs and their fragmentation pathways. The chemical structures and the corresponding peak numbers 1 to 5 represent signals for N-nornuciferine, O-nornuciferine, anonaine, nuciferine, and Roemerine, respectively. Figures are modified from Luo et al. [65] and Chen et al. [23].
-

Figure 7.
Analytical mass spectra of lotus leaf BIAs and their fragmentation pathways. The chemical structures and the corresponding peak numbers 6 to 8 represent signals for liensinine, isoliensinine, and neferine, respectively. Figures are modified from Chen et al. [64], Deng et al. [11], and Lai et al. [66].
-
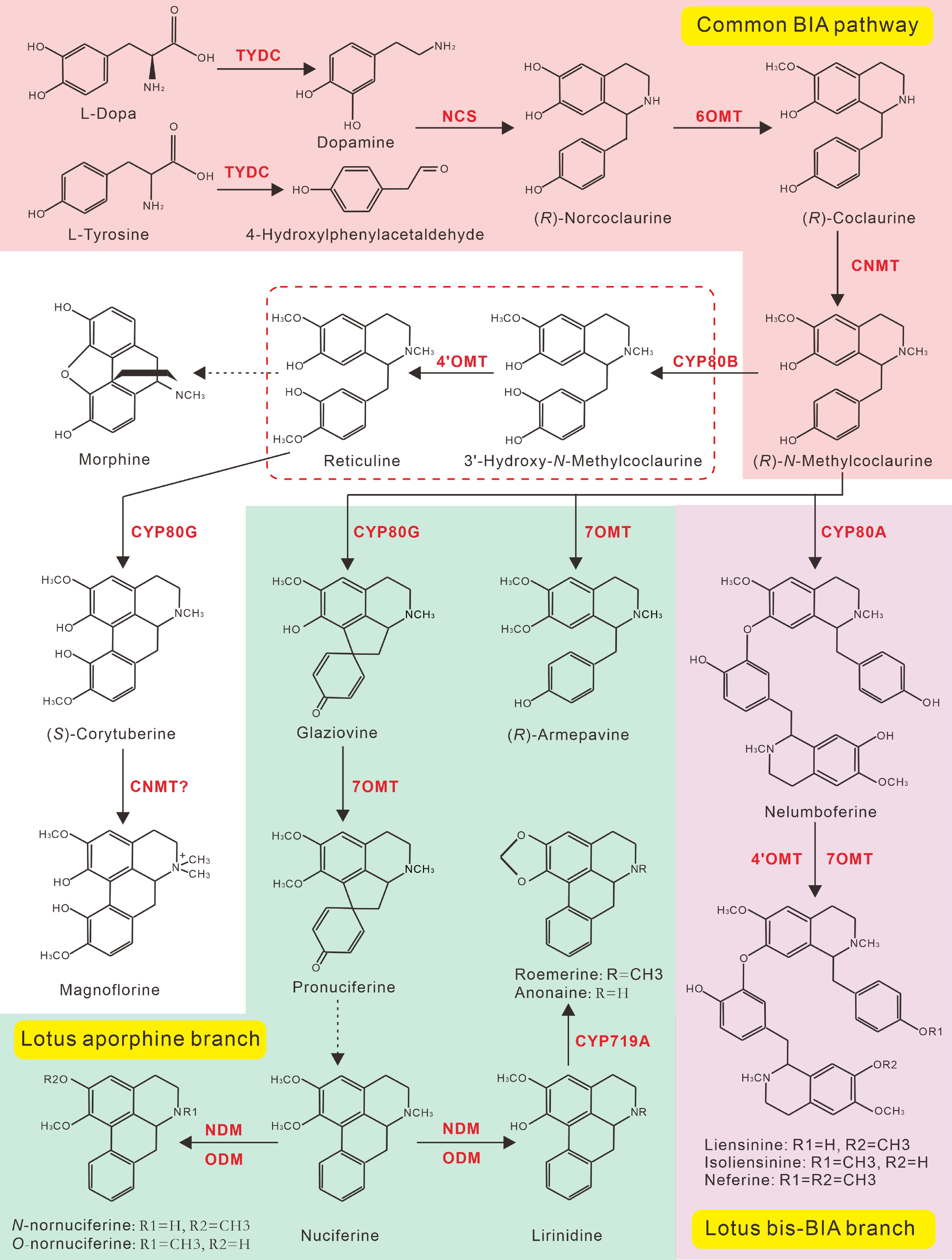
Figure 8.
Proposed BIAs biosynthetic pathway in lotus. Steps marked with red, green, and purple background represent the common reactions for the biosynthesis of (R)-N-Methylcoclaurine, the lotus aporphine branch, and the bis-BIA biosynthesis branch, respectively. All biosynthetic enzymes are shown in red. The (R)-N-Methylcoclaurine is the branch point for aporphine and bis-BIA biosynthesis in lotus. Dotted arrows indicate multiple enzymatic or unknown steps.
-
No. Alkaloid Formula Enantiomer Organ Reference 1-BENZYLISOQUINOLINE 1 Lotusine C19H24NO3+ E, S, F [25] 2 Methyl lotusine L 3 Armepavine C19H23NO3 (−)-R and (+)-S S, F, L [21] 4 Coclaurine C17H19NO3 (+)-R S, L [26] 5 N-norarmepavine C18H21NO3 (+)-R L [27] 6 N-methylisococlaurine C18H21NO3 L [16,28] 7 N-methylcoclaurine C18H21NO3 (−)-R L, S [16] 8 Isococlaurine C19H24NO3+ L 9 Methylhigenamine S [17] 10 Norcoclaurine-6-O-glucoside S 11 Norcoclaurine C16H17NO3 (+)-R and (+)-S S, L [26,29] 12 Argemexirine S, L 13 6-demethy-4-methyl-N-methylcoclaurine C18H21NO3 S [30] 14 Nor-O-methylarmepavine C20H25NO3 S [30] 15 4’-N-methylcoclaurine C19H23NO3 L, S [17] 16 4’-methyl coclaurine L, S [17] 17 Bromo methyl armepavine L, S [17] 18 Methoxymethy lisoquinoline L, S [17] 19 Higenamine P [31,32] 20 Higenamine glucoside P [33] APORPHINE 21 Nuciferine C19H21NO2 (−)-R S, F, L [34,35] 22 N-nornuciferine C18H19NO2 (−)-R S, L [26] 23 Roemerine C18H17NO2 (−)-R S, F, L [16,26,32] 24 O-nornuciferine C18H19NO2 (−)-R S, F, L [32,36] 25 Anonaine C17H15NO2 (−)-R S, L [16,26] 26 Lirinidine C18H19NO2 (−)-R S, L [37] 27 Nuciferine-N-Methanol F 28 Nuciferine-N-Acetyl F 29 Anonaine-N-Acetyl F 30 Caaverine C17H17NO2 (−)-R S, L [20,38] 31 Oxidation-nuciferine S, L, F [19,30] 32 Asimilobine C17H17NO2 (−)-R S, F, L [17,20] 33 Methyl asimilobine S, L [17] 34 N-methyl asimilobine S, L [16,39] 35 Roemerine-N-oxide S, L [16,40] 36 N-methyl asimilobine-N-oxide S, L, F [16,19] 37 Nuciferine-N-oxide S, L, F [16,19] 38 Dehydroanonaine C17H13NO2 L [16] 39 Dehydronuciferine C19H19NO2 L [16] 40 Dehydroaporphine C18H15NO2 L [41] 41 Nelumnucine S, L 42 Dehydroroemerine L [16] 43 Liriodenine C17H9NO3 L [16,40] 44 7-hydroxydehy dronuciferine C19H19NO3 L [16] 45 Pronuciferine C19H21NO3 (+)-R and (−)-S S, F, L [16,26,40] 46 Glaziovine S 47 Lysicamine C18H13NO3 L [38,42] 48 Cepharadione L [38] BISBENZYLISOQUINOLINE 49 Neferine C38H44N2O6 1R, 1'S S, F, E [17,31] 50 Liensinine C37H42N2O6 1R, 1'R S, F, E [35] 51 Isoliensinine 1R, 1'S S, F, E [25] 52 N-norisoliensinine C36H40N2O6 S, F, E [25] 53 6-hydroxynorisoliensinine C36H40N2O6 S, F, E 54 Methyl neferine S, E [10,17] 55 Nelumboferine C36H40N2O6 S, E [10,17] 56 Negferine C38H44N2O6 L [17,26] 57 Nelumborine F [17] 58 Dauricine S, F [43] TRIBENZYLISOQUINOLINE 59 Neoliensinine C63H70N3O10 1R, 1'S, 1''R E [44] L, Leaf; E, embryo; F, flowers; S, seeds; R, rhizome; LS: leaf sap; NS, not specified. -
Extraction method Content (mg/g dry weight)a Total 1 2 3 4 5 6 7 8 9 10 Methanol, reflux 1.76 (100) 1.75 (100) 0.07 (100) 0.63 (100) 0.69 (100) 0.83 (100) 1.45 (100) 5.73 (100) 1.30 (100) 0.75 (100) 14.96 (100) 50% Methanol, reflux 1.09 (62) 1.35 (77) 0.05 (71) 0.50 (79) 0.61 (88) 0.78 (94) 1.35 (93) 3.79 (66) 0.94 (73) 0.56 (75) 11.02 (74) H2O, reflux 0.24 (14) 0.35 (20) nd.b 0.21 (33) 0.18 (26) 0.38 (45) 0.78 (54) 2.57 (45) 0.66 (51) 0.29 (38) 5.66 (38) Methanol, sonication 0.88 (50) 1.11 (64) 0.03 (44) 0.39 (62) 0.33 (48) 0.47 (56) 0.97 (67) 2.77 (48) 0.70 (54) 0.42 (56) 8.07 (54) 50% Methanol, sonication 0.98 (56) 1.27 (73) 0.04 (58) 0.49 (78) 0.47 (69) 0.80 (96) 1.38 (95) 3.93 (69) 0.97 (75) 0.59 (79) 10.92 (73) H2O, sonication 0.14 (8) 0.21 (12) nd. 0.12 (20) 0.08 (11) 0.25 (30) 0.53 (37) 1.91 (33) 0.48 (37) 0.19 (26) 3.91 (26) 1. nuciferine; 2. N-nornuciferine; 3. N-methylasimilobine; 4. Asimilobine; 5. Pronuciferine; 6. Armepavine; 7. norarmepavine; 8. N-methylcoclaurine; 9. coclaurine; 10. Norjuziphine. a. relative value (%) against the content obtained by methanol under reflux is given in parentheses. b. less than the quantitation limit. Table 2.
Extraction efficiency of alkaloids from lotus flower.
-
Solvent systems Mix ratios (v/v) Target BIAs Reference Light petroleum (60–90 °C)–ethyl acetate–tetrachloromethane–chloroform–methanol–water 1:1:4:4:6:2 Small scale
Plumule BIAs[59] Ethyl acetate–tetrachloromethane–chloroform–methanol–water 1:6:4:1 Small scale
Plumule BIAs[59] n-hexane–ethyl acetate–methanol–water 5:8:4:5
0.5% NH4OHPlumule BIAs [60] n-hexane–ethyl acetate–methanol–water 5:5:2:8
10 mM triethylamine
5 mM HClPlumule BIAs [57] Diethyl ether – Na2HPO4/NaH2PO4 (pH = 7.2 – 7.5) 1:1 Plumule BIAs [62] Petroleum ether (60–90 °C)–ethyl acetate–methanol–water 5:5:2:8
10 mM triethylamine
5 mM HClLeaf BIAs [61] n-hexane-ethyl acetate-methanol-water-[C4mim][PF6] 5:2:2:8:0.1
10 mM triethylamine
3 mM HClWhole plant BIAs [35] Table 3.
Major two-phase solvent systems developed for preparative separation of lotus BIAs through counter-current chromatography (CCC) techniques.
-
Peaks TRa (min) Molecular weight m/z
[M + H]+Major fragment ions Alkaloids 1* 7.56 281 282 251/219 N-nornuciferine 2* 9.03 281 282 265/250 O-nornuciferine 3* 10.17 265 266 249/219 Anonaine 4* 12.43 295 296 265/250 Nuciferine 5* 13.39 279 280 249 Roemerine 6∆ 6.61 610 611 503/283/206 Liensinine 7∆ 9.47 610 611 489/297/192 Isoliensinine 8∆ 17.72 624 625 503/297/206 Neferine * The retention time (TRa (min) of peaks 1–5 as obtained by Chen et al.[23].
∆ The retention time of peaks 6–9 as reported by Chen et al.[63].Table 4.
Identification of major lotus leaf and plumule alkaloids and their HPLC-MS/MS ion characteristics.
Figures
(8)
Tables
(4)