-

Figure 1.
Carya illinoinensis cv. Xinxuan-4. (a) Male flower. (b) Female flower. (c) Fruits on the tree. (d) Fruit without husk, scale bar = 2 cm.
-

Figure 2.
Circular representation of the cp genome of C. illinoinensis cv. Xinxuan-4.
-
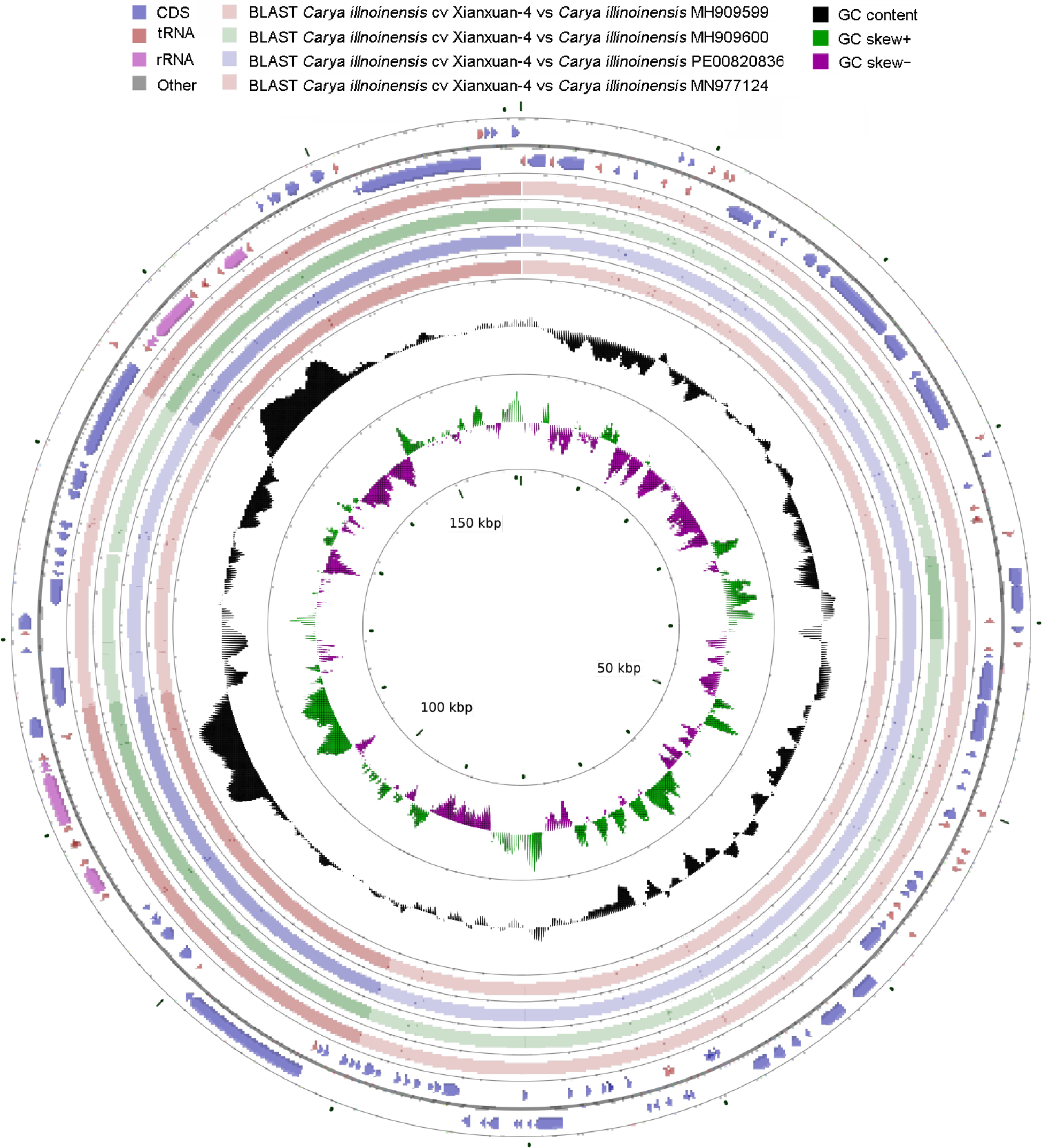
Figure 3.
GC content of the C. illinoinensis cv. Xinxuan-4 cp genome.
-

Figure 4.
Codon usage frequency of the C. illinoinensis cv. Xinxuan-4 cp genome.
-

Figure 5.
Size and type of the long repeats located in the C. illinoinensis cv. Xinxuan-4 cp genome.
-

Figure 6.
SSRs distribution in the cp genomes of the C. illinoinensis cv. Xinxuan-4. (a) Type and number of SSRs. (b) Type and number of SSR repeats. (c) Number of SSRs in various location. (d) Number of SSRs in three locations.
-

Figure 7.
Comparing the LSC, SSC, and IR locations of five selected cp genomes in the Carya.
-
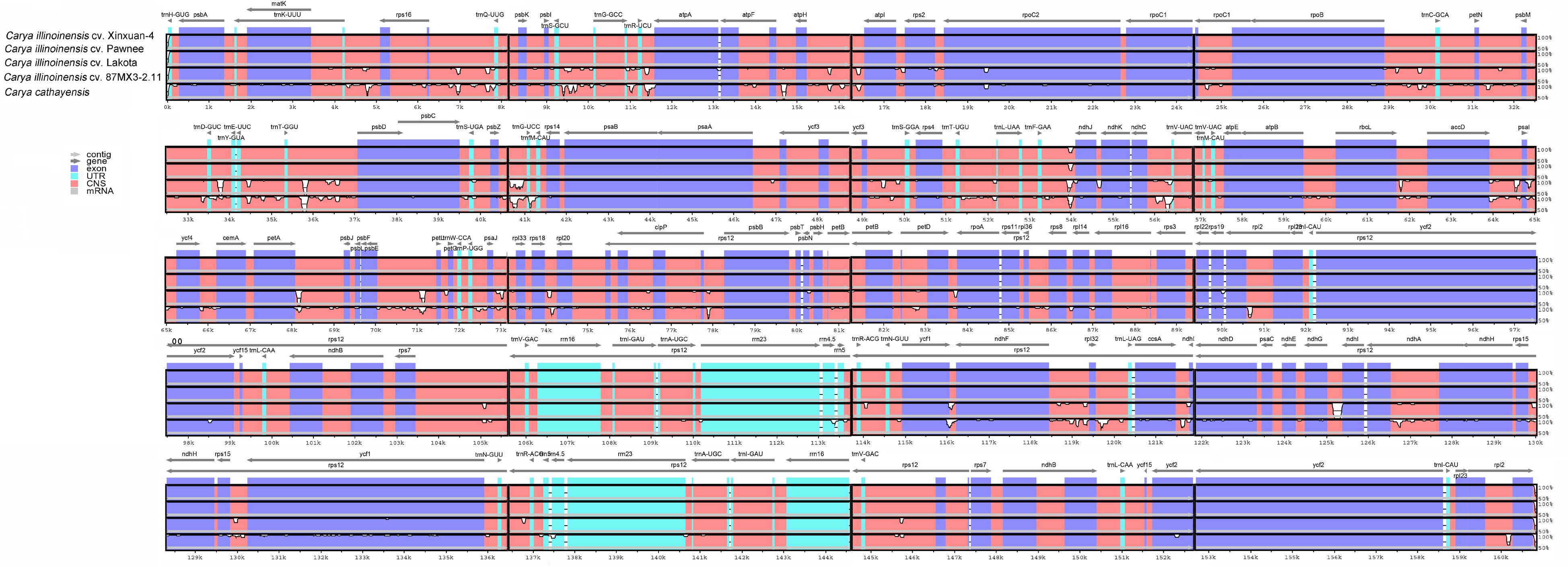
Figure 8.
Sequence identity plot comparing the cp genomes among Carya with C. illinoinensis cv. Xinxuan-4 set as a reference.
-
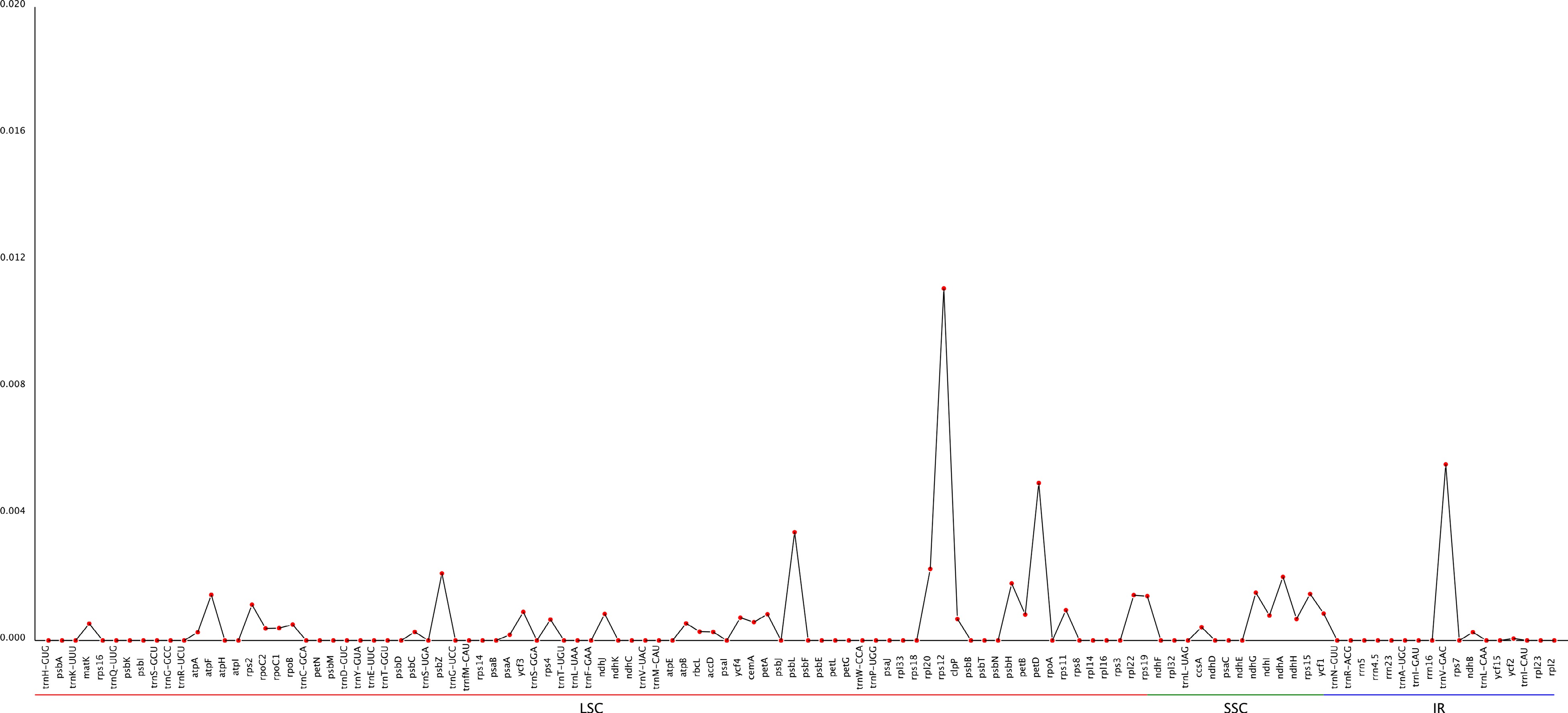
Figure 9.
Sliding window analysis of the cp genome for nucleotide diversity (pi) of three species in Carya.
-
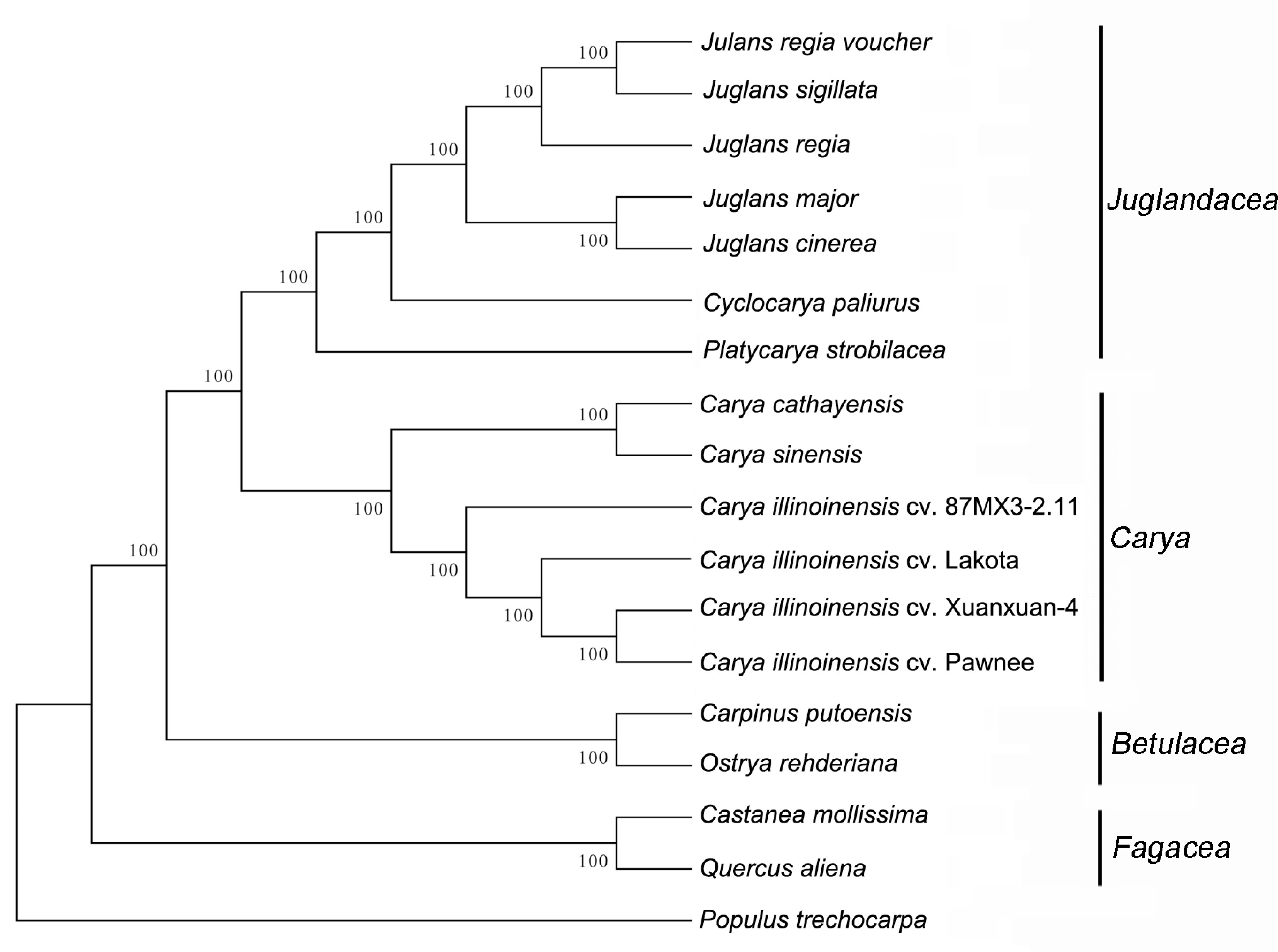
Figure 10.
Phylogenetic tree of 18 related species based on the whole cp genome.
-
Species Xinxuan-4 Pawnee 87MX3-2.11 Lakota Wichita Cathayensis Genome size (bp) 160,819 160,819 160,545 160,819 160,532 160,825 LSC size (bp) 90,022 90,022 89,933 90,041 89,799 90,115 SSC size (bp) 18,791 18,791 18,576 18,790 18,751 18,760 IR size (bp) 26,003 26,003 26,018 25,994 25,991 25,975 Total genes 132 131 124 123 128 129 Protein-coding genes 87(8) 86(7) 84(6) 83(6) 83(6) 84 tRNAs 37(7) 37(8) 32(6) 32(6) 37(8) 37(7) rRNAs 8(4) 8(4) 8(4) 8(4) 8(4) 8(4) GC content (%) 36.1 36.1 36.2 36.1 36.2 36.1 Table 1.
Features of the cp genomes of C. illinoinensis cv. Xinxuan-4 and five related materials.
-
Category Gene group Photosynthesis Subunits of photosystem I psaA, psaB, psaC, psaI, psaJ Subunits of photosystem II psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ Subunits of NADH ndhAb, ndhBab, ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK Cytochrome b/f complex petA, petBb, petDb, petG, petL, petN Subunits of ATP synthase atpA, atpB, atpE, atpFb, atpH, atpI Large subunit of rubisco rbcL Self-replication Large ribosomal subunit rpl14, rpl16b, rpl2ac, rpl20, rpl22, rpl23a, rpl32, rpl33, rpl36 Small ribosomal subunit rps11, rps12ab, rps14, rps15, rps16b, rps18, rps19, rps2, rps3, rps4, rps7a, rps8 Subunits of RNA polymerase rpoA, rpoB, rpoC1b , rpoC2 Ribosomal RNAs rrn16a, rrn23a, rrn4.5a, rrn5a Transfer RNAs trnA-UGCab, trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnG-GCCb, trnG-UCC, trnH-GUG, trnI-CAUa, trnI-GAUab, trnK-UUUb, trnL-CAAa, trnL-UAAb, trnL-UAG, trnM-CAU, trnN-GUUa, trnP-UGG, trnQ-UUG, trnR-ACGa, trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU, trnT-UGU, trnV-GACa, trnV-UACb, trnW-CCA, trnY-GUA, trnfM-CAU Other genes Maturase matK Protease clpPc Envelope membrane protein cemA Acetyl-CoA carboxylase accD c-type cytochrome synthesis gene ccsA Unknown function genes Conserved open reading frames ycf1a, ycf15a, ycf2a, ycf3c, ycf4 a Gene with two copies, b Gene with one intron, c Gene with two introns. Table 2.
Annotated genes in the cp genome of C. illinoinensis cv. Xinxuan-4.
-
Gene group Different gene Xinxuan4 Pawnee 87MX3-2.11 Lakota Cathayensis Protein-coding genes rps12 2 2 1 0 2 rps16 1 1 0 0 1 psbZ 1 1 1 1 0 lhbA 0 0 0 0 1 ycf15 2 2 2 2 0 tRNA genes trnA-UGC 2 2 2 2 0 trnG-UCC 1 0 0 0 0 trnI-GAU 2 2 0 0 0 trnL-UAA 1 1 0 0 1 trnV-UAC 1 1 0 0 1 rna-UGC 0 0 0 0 2 Table 3.
Different genes and gene copies in the cp genomes among five Carya materials.
Figures
(10)
Tables
(3)