-
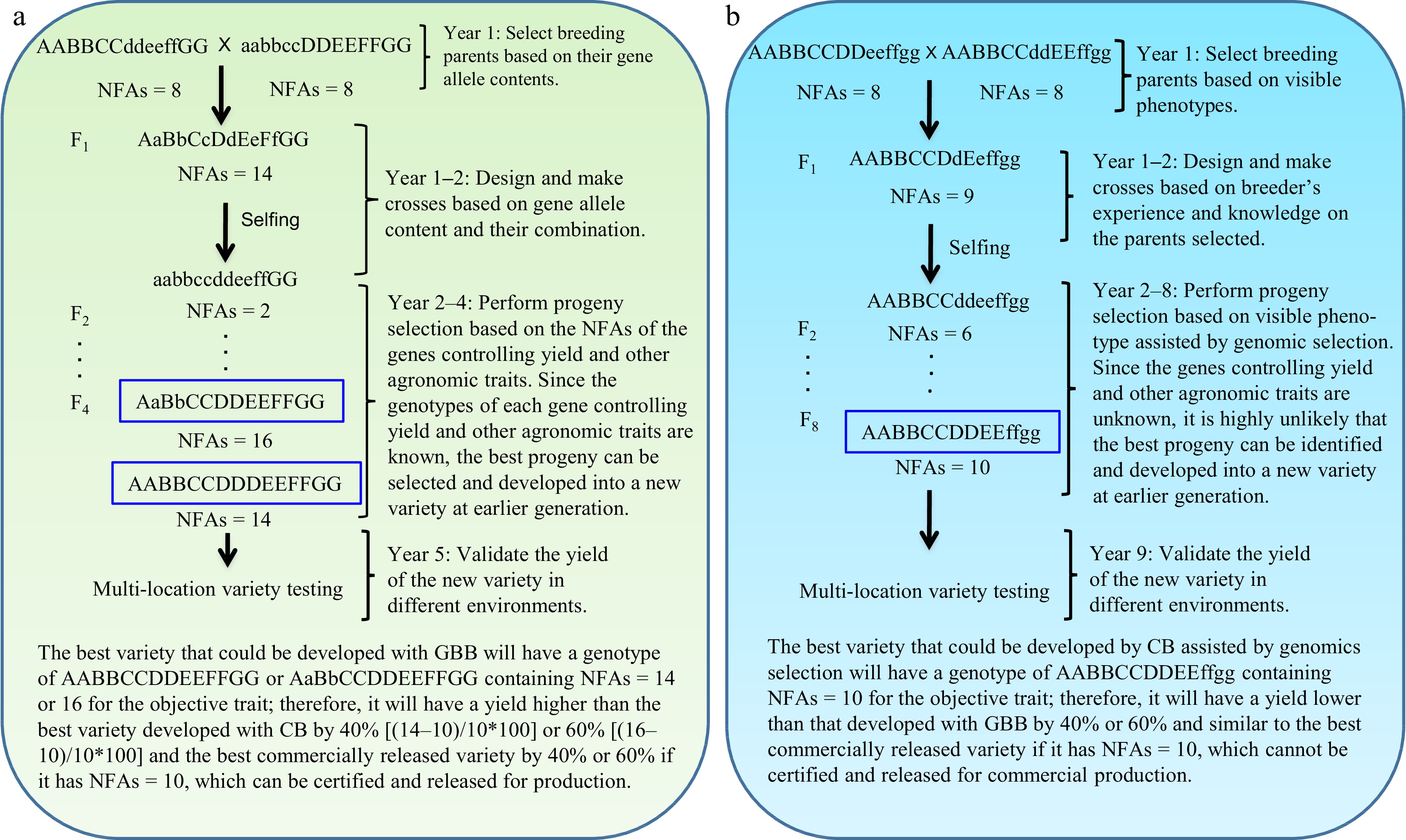
Figure 1.
Gene-based breeding (GBB) for pure-line varieties using (a) the total NFAs of the genes controlling breeding objective trait (yield) vs (b) the current breeding (CB) assisted by genomic selection. NFAs of the genes controlling yield are used for parent selection, crossing design, and progeny selection. Where a GBB variety is released for commercial production will be determined by multi-location variety testing. Assuming that yield is controlled by seven genes, with AA and BB having over-dominant effects (NFAs = 3 for Aa or Bb); CC, EE, and GG having additive effects (NFAs = 1 for Cc, Ee, or Gg); and DD and FF having complete dominant effects (NFAs = 2 for Dd or Ff) on yield. The total NFAs of the genes in an individual plant is the sum of the NFAs of individual genes for the plant. Therefore, the parents selected, with each having NFAs = 8, for GBB and CB, have similar phenotypes for the objective trait, but they have different contents of the genes controlling the objective trait. As yield is controlled by more than one thousand genes[13], it is impossible for breeders to identify the most desirable breeding lines as breeding parents and optimally combine most, if not all, of them into a new variety with CB. GBB can do so because it allows identification of the breeding parents best meeting the breeding objectives and allows optimal crossing design that can maximally combine the favorable alleles and heterosis genotypes into a new variety. The current best commercial variety is assumed to have NFAs = 10.
-
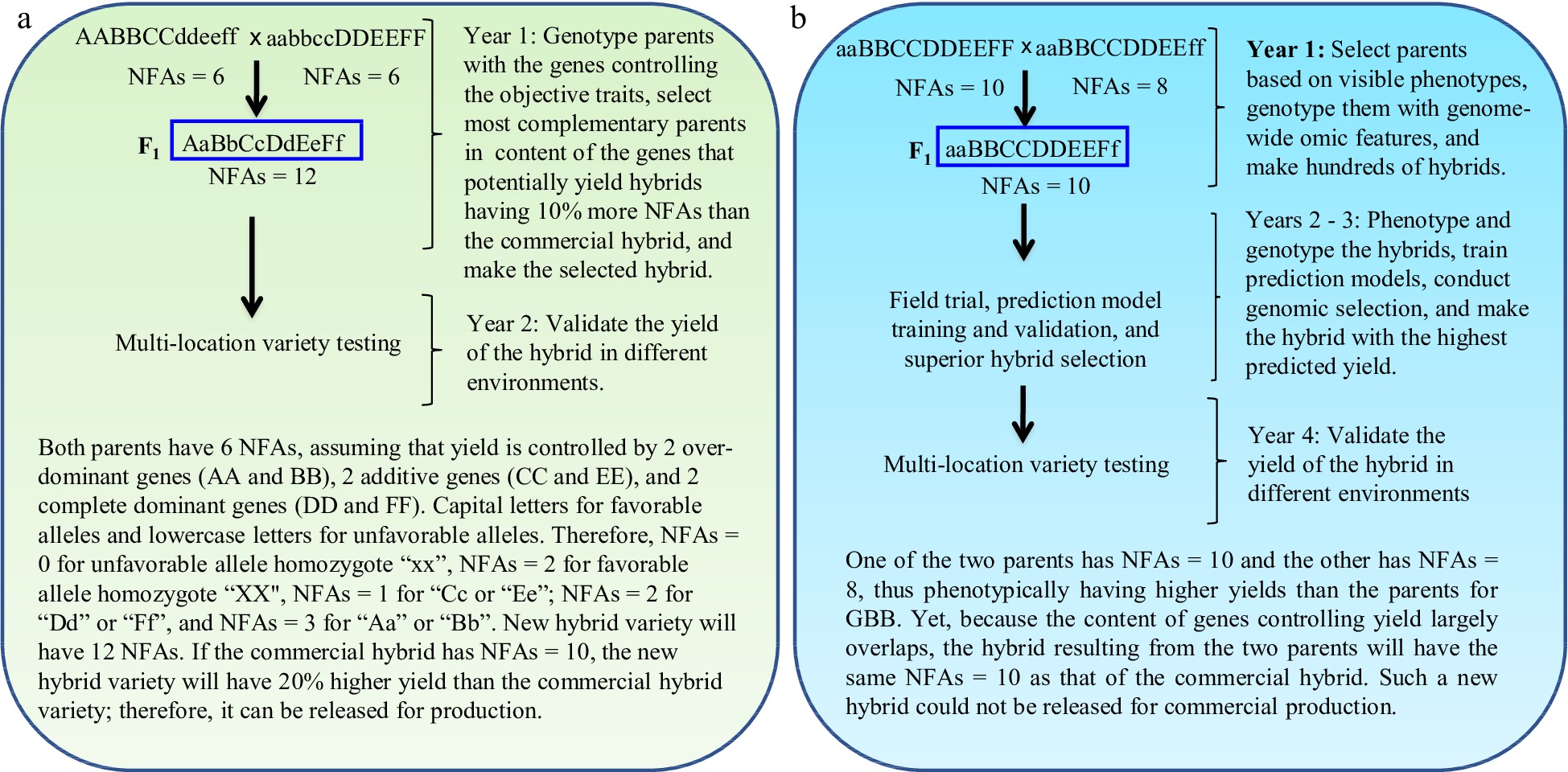
Figure 2.
Gene-based breeding (GBB) for hybrid varieties using (a) the total NFAs of the genes controlling breeding objective trait (yield) vs (b) the current breeding (CB) assisted by genomic selection. The NFAs of the genes controlling the objective trait are used for parent selection, crossing design, and progeny selection. Simplifying for example that yield is controlled by six genes, with capital letters for favorable alleles and lowercase letters for unfavorable alleles. The total NFAs of the genes in a hybrid is the sum of the NFAs of individual genes for the hybrid. The parents selected for GBB, with each having NFAs = 6 for the objective trait, have lower grain yields than those selected for CB, with each having NFAs = 8 or 10. As yield is controlled by more than one thousand genes[13,14], it is impossible for breeders to identify the most desirable inbred lines as breeding parents and optimally combine most, if not all, of them into a new hybrid variety with CB. GBB can do so because it allows identification of the breeding inbred line parents best meeting the breeding objectives and allows optimal crossing design that can maximally combine the favorable alleles and heterosis genotypes into a new hybrid variety. The current best commercial hybrid variety is assumed to have NFAs = 10.
-

Figure 3.
Prediction accuracies of agronomic traits using genic datasets of the genes controlling breeding objective traits for GBB. (a) Prediction accuracies of cotton fiber length using the SNPs/InDels contained in 226 GFL (Gossypium fiber length) genes[12]. The 226 SNPs/InDels were selected from the 740 SNPs/InDels, with one SNP per GFL gene. (b) Prediction accuracies of cotton fiber length using the total NFAs of the 226 GFL genes versus those predicted with 3,000 genome-wide random SNPs[15]. (c) Correlation of predicted cotton fiber lengths between the NFAs and the SNPs of the 226 GFL genes[15]. (d) Correlation of predicted cotton fiber lengths between the NFAs and the transcript expressions of the 226 GFL genes[15]. (e) Prediction accuracies of maize inbred line grain yields using the expressions of the transcripts of the ZmINGY (Zea mays inbred grain yield) genes responsible for grain yield[13,27]. (f) Prediction accuracies of maize hybrid grain yields from their parents using the expressions of the transcripts of the ZmF1GY (Zea mays F1 hybrid grain yield) genes responsible for hybrid grain yield vs those with the expressions of 122,029 genome-wide random transcripts[14]. For the abbreviations of prediction models, see Liu et al.[12,15] & Zhang et al.[13,14].
-

Figure 4.
Genetic potential of current cotton varieties or advanced breeding lines having a fiber length of 33.8 mm predicted using the total NFAs of the 226 GFL genes contained in cotton advanced breeding plants or lines[16]. (a) Training linear and non-linear models using the fiber lengths of 198 advanced breeding lines determined by multiple replicated field trials and the total NFAs of their 226 GFL genes controlling fiber length. (b) Predicting the genetic potential of the current cotton variety or advanced breeding line with the longest fiber length using the trained linear and non-linear models, respectively. The current cotton variety or advanced breeding line with the fiber lengths (33.8 mm) of the current best cotton varieties only contained 52% of the 728 total NFAs of the 226 GFL genes and that they could potentially be further improved by up to 118% for the linear model or 73% for the non-linear model, if all 728 NFAs of the 226 GFL genes are incorporated into a new variety through GBB.
-
Consistency of GBB with PS ZmINGY genic datasets I II III I + II I + III II + III I + II + III Field trials, Halfway, Texas, 2010 40.0% 50.0% 66.7% 100.0% 66.7% 100.0% 100.0% Field Trials, College Station, Texas, 2010 41.2% 33.3% 55.6% 80.0% 100.0% 100.0% 100.0% I. Number of favorable alleles (NFAs) of 27 SNP/InDel-containing ZmINGY genes; II. SNPs/InDels of the 27 SNP/InDel-containing ZmINGY genes; III. The transcript expressions of the 150 key ZmINGY genes. Note that when the grain yields of the plants predicted with two or all three genic datasets of the ZmINGY genes were jointly used for progeny selection, the top 10% plants selected with the highest grain yields predicted with the genes were consistent up to 100% with those selected with the highest grain yields determined by replicated field trials. Halfway, Texas and College Station, Texas represent two different agricultural ecosystems and climate zones in the USA. Table 1.
Progeny selection of gene-based breeding (GBB) for top 10% plants with the highest grain yields predicted with ZmINGY genes vs phenotypic selection (PS) of conventional breeding for top 10% plants with the highest grain yields determined by replicated field trials for inbred line breeding in maize[13].
-
Statistical analysis (ANOVA and LSD) Effect Genotype of a gene AA Aa aa AA > Aa > aa Additive 2 1 0 Aa = AA > aa complete dominant 2 2 0 Aa > AA > aa Over-dominant 2 3 0 Allele 'A' is the favorable allele over allele 'a', when 'AA' is larger than 'aa' (p ≤ 0.05). The total NFAs of all genes controlling an agronomical trait is calculated with the following formula: y = ∑xi, where y is the total NFAs of the genes controlling an agronomical trait, x is the NFAs of individual genes controlling the trait, which is 0, 1, 2, or 3, in individual 'i'. Table 2.
Statistics of NFAs of each gene and total NFAs of all genes controlling an agronomical trait[15].
Figures
(4)
Tables
(2)