-
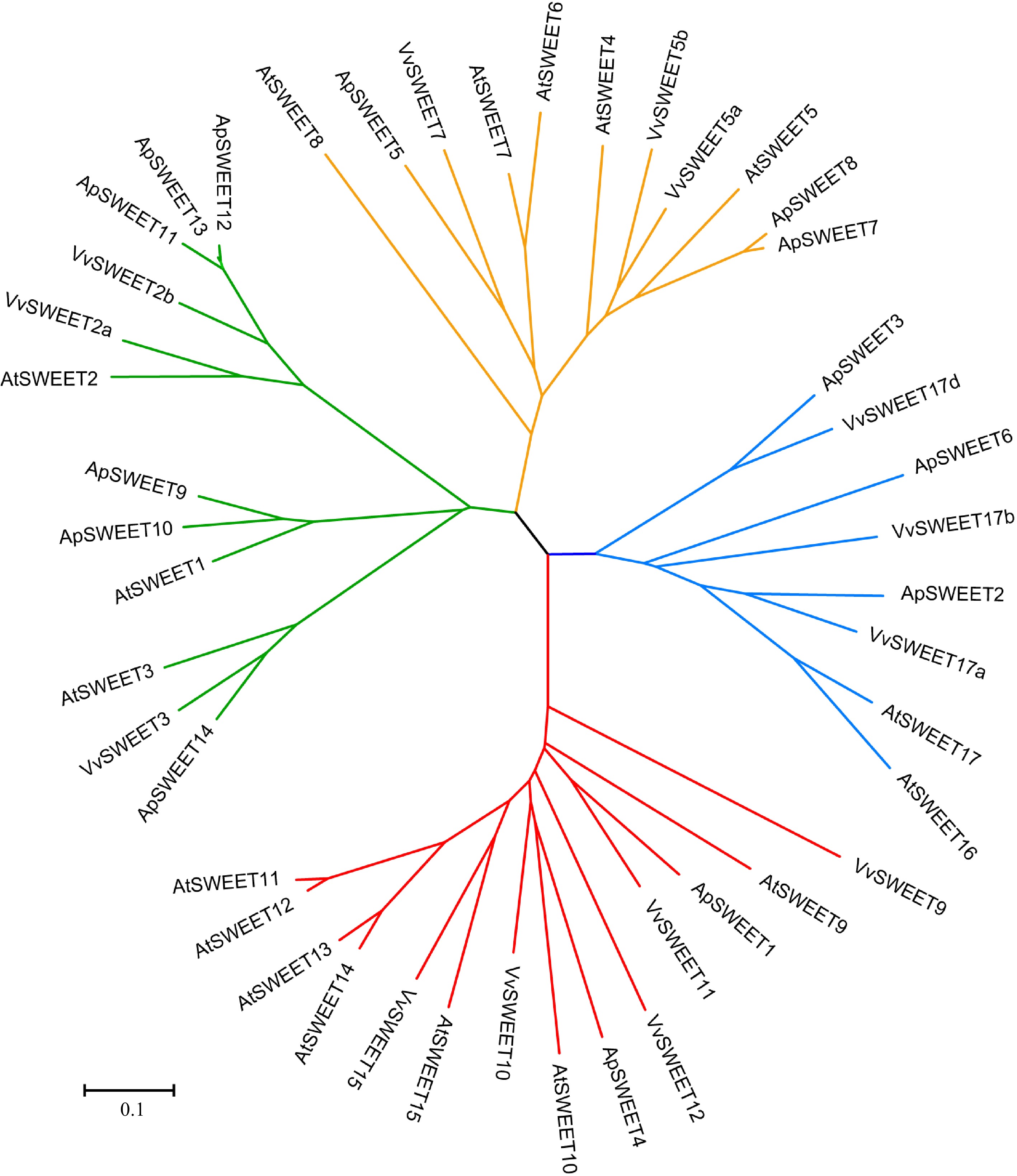
Figure 1.
Phylogenetic analysis of the ApSWEETs from A. polygama, Arabidopsis thaliana, and Vitis vinifera. The Neighbor-joining tree was drawn using MEGA7.0 with 1,000 bootstraps. The roman numbers (I–IV) labeled with various colors indicate different clades: green – Clade I, orange – Clade II, red – Clade III, blue – Clade IV.
-
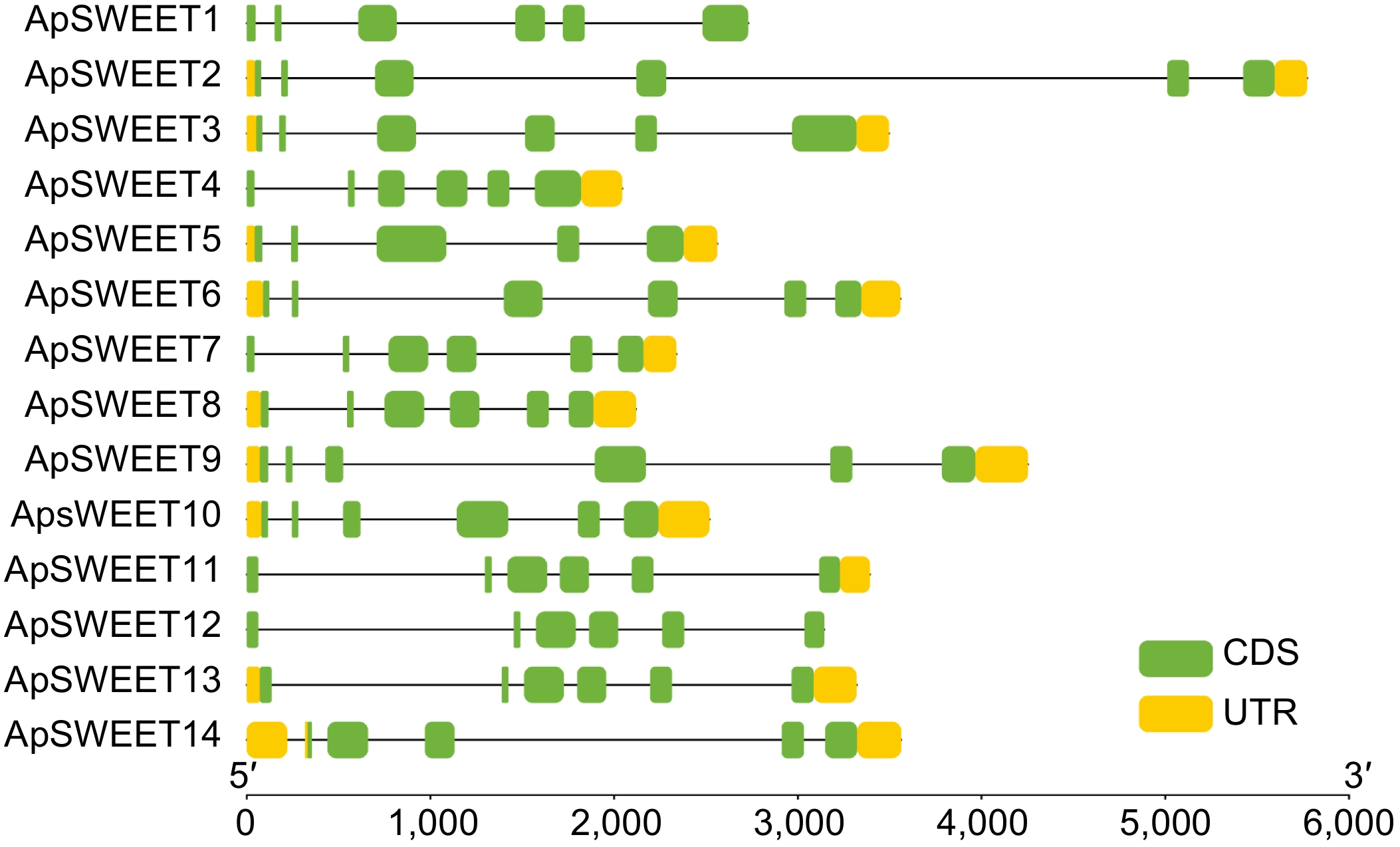
Figure 2.
ApSWEET gene structure of A. polygama.
-
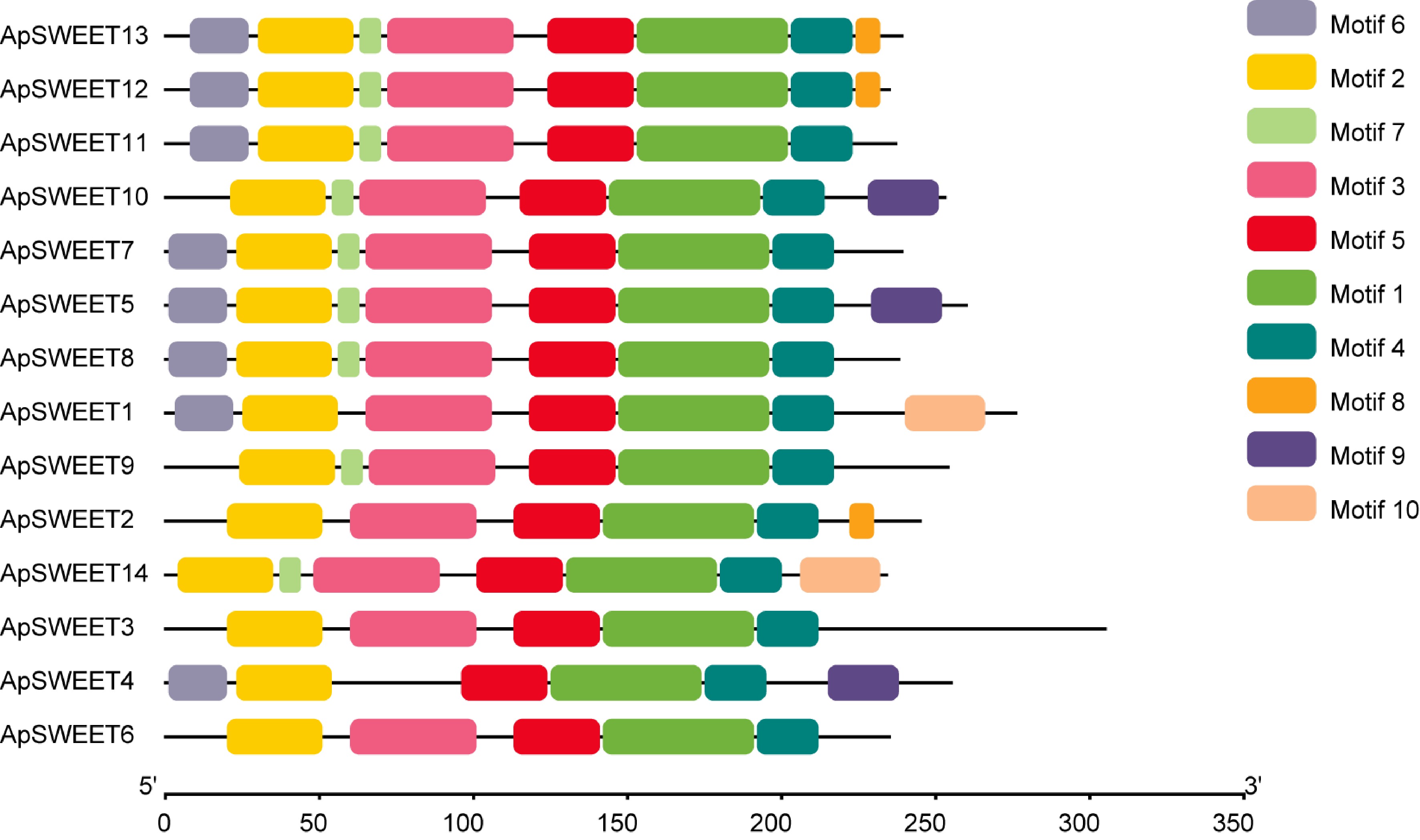
Figure 3.
The conserved motif analyses of ApSWEETs proteins.
-
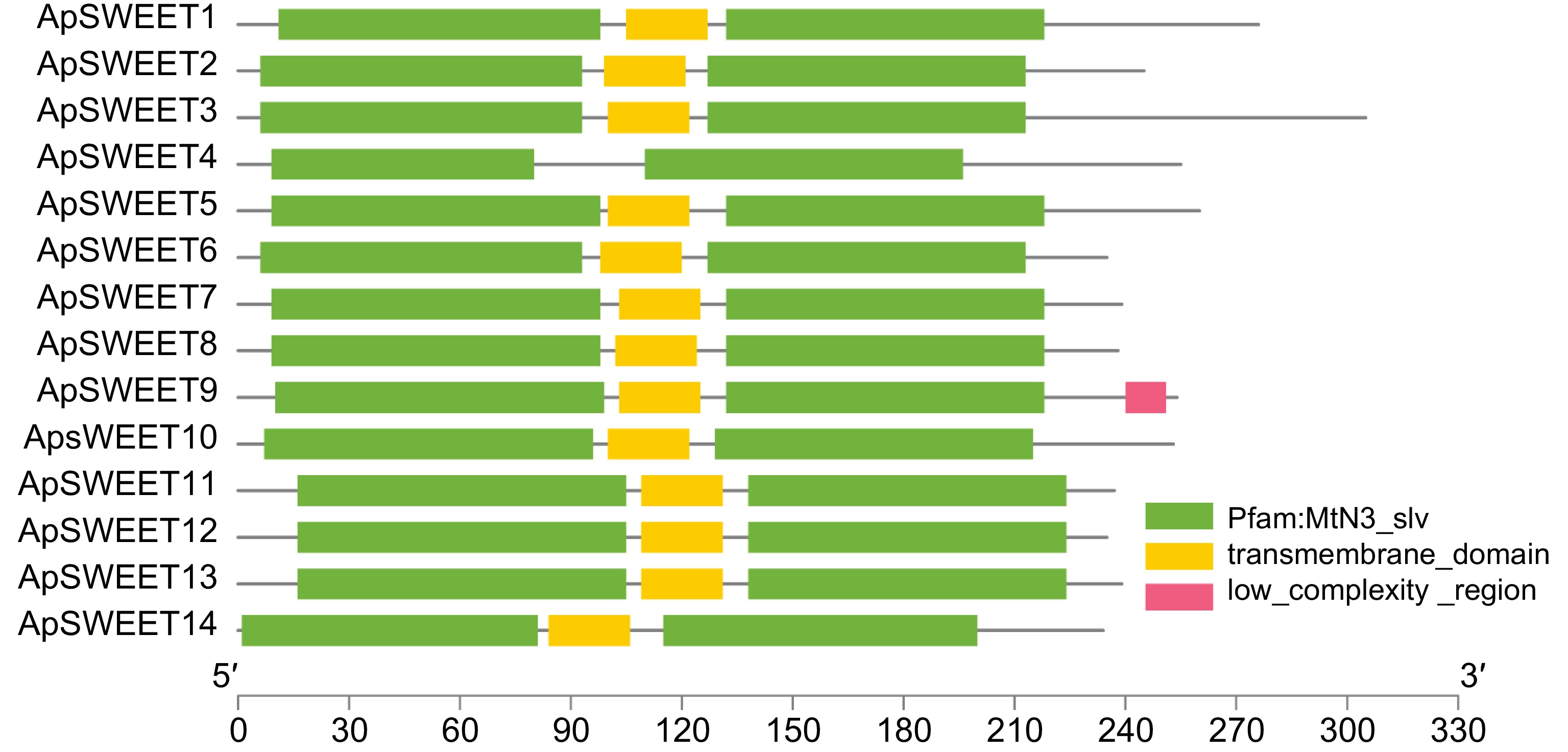
Figure 4.
Conserved structural domains of ApSWEETs.
-
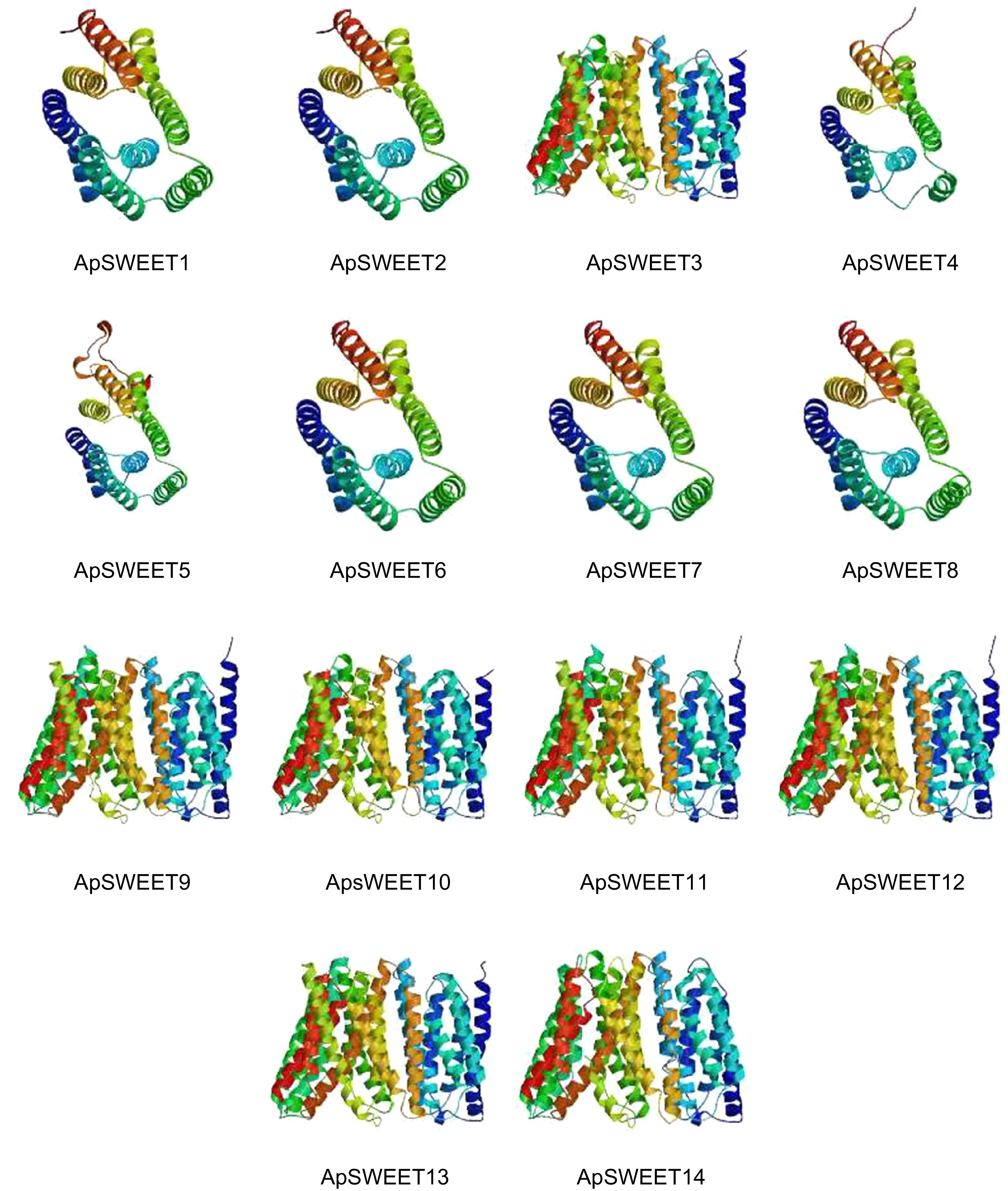
Figure 5.
Single protein structure of ApSWEETs.
-
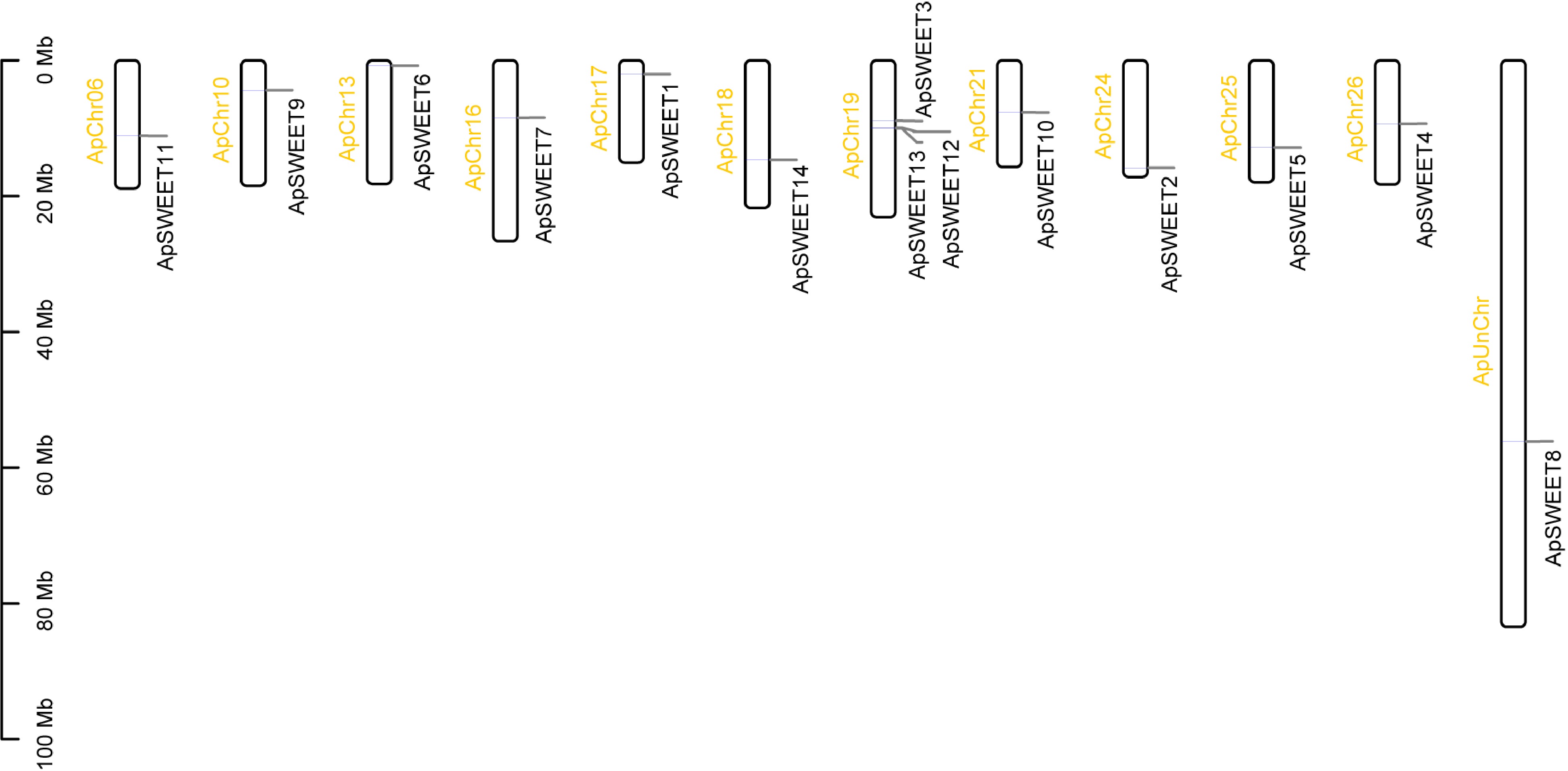
Figure 6.
Chromosome location of ApSWEET gene family.
-
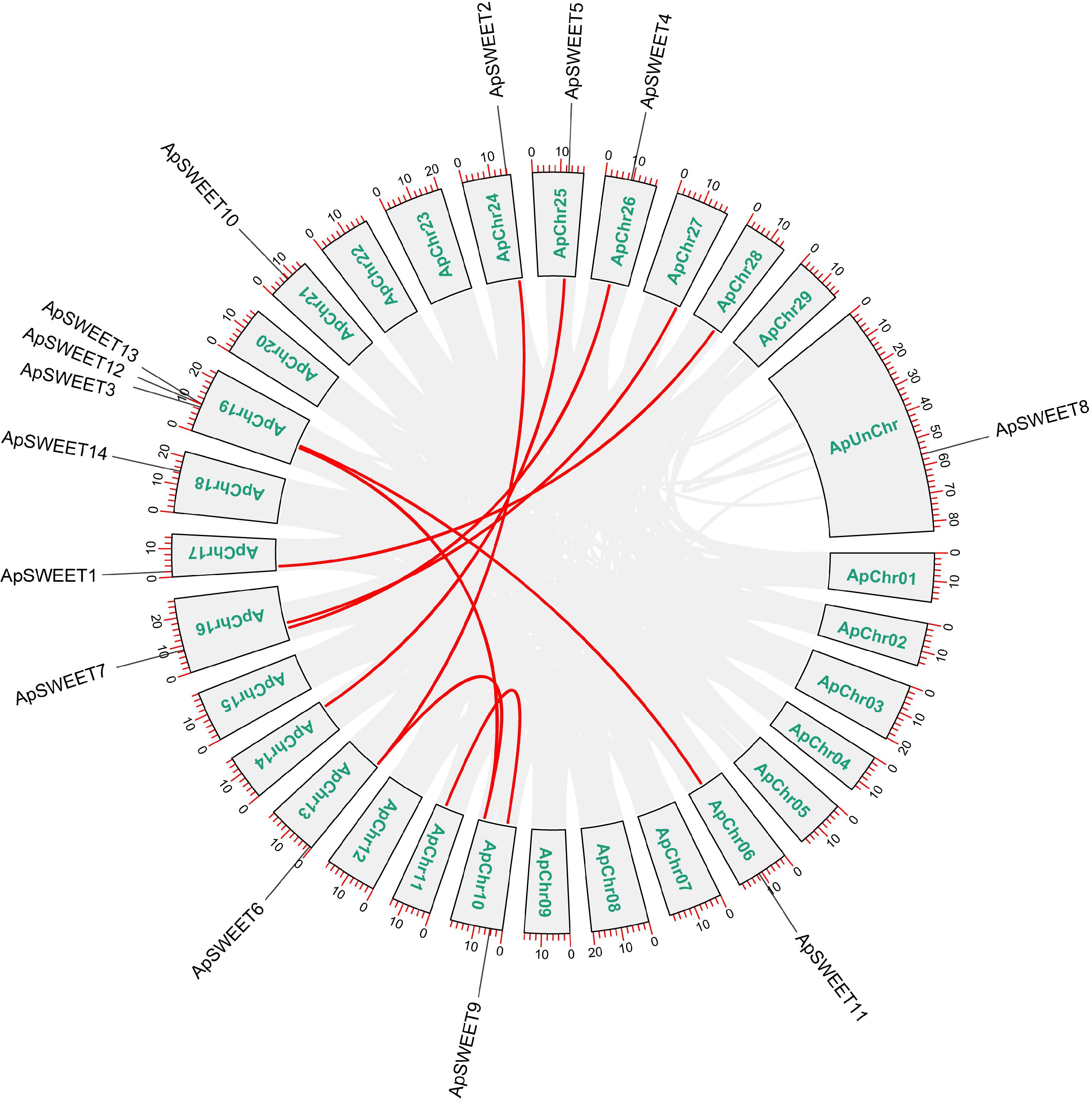
Figure 7.
Collinearity analysis of the ApSWEET gene family.
-
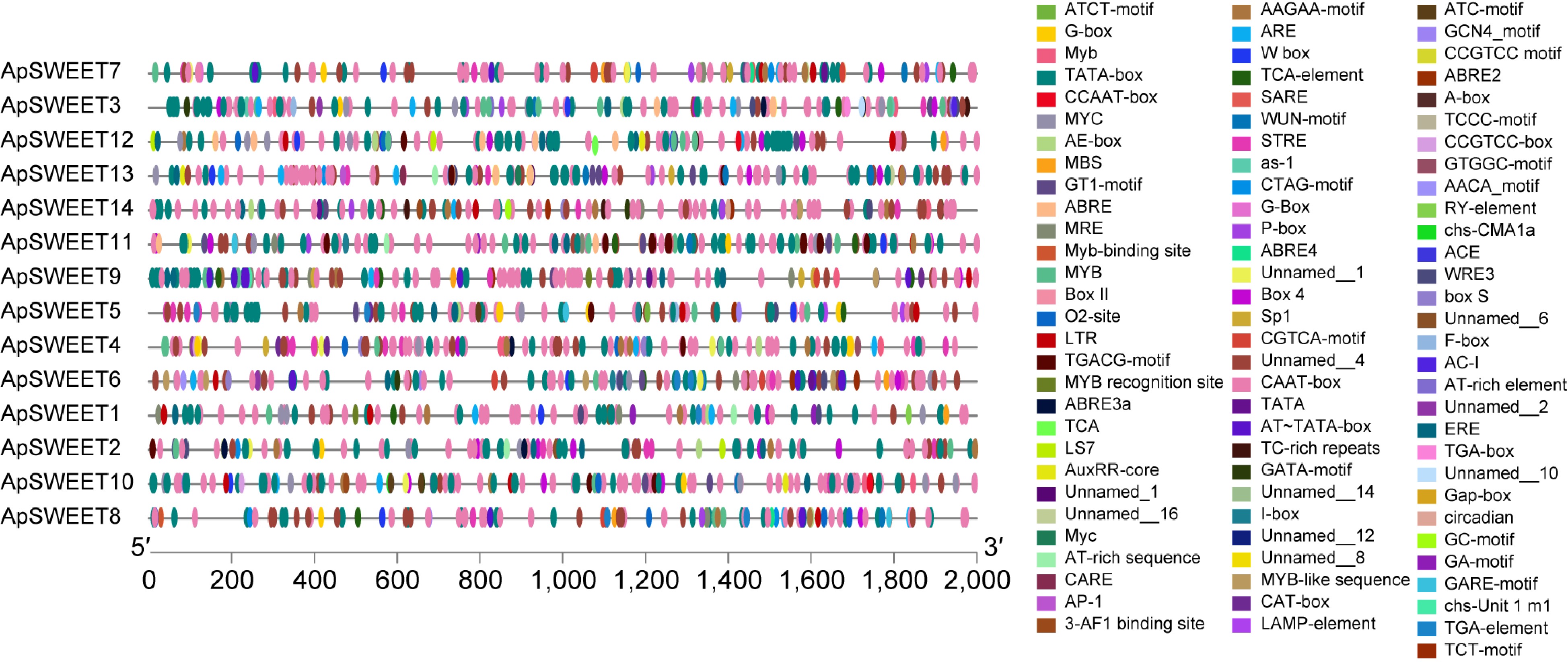
Figure 8.
The cis-elements in the promoter sequences of ApSWEETs gene in A. polygama.
-
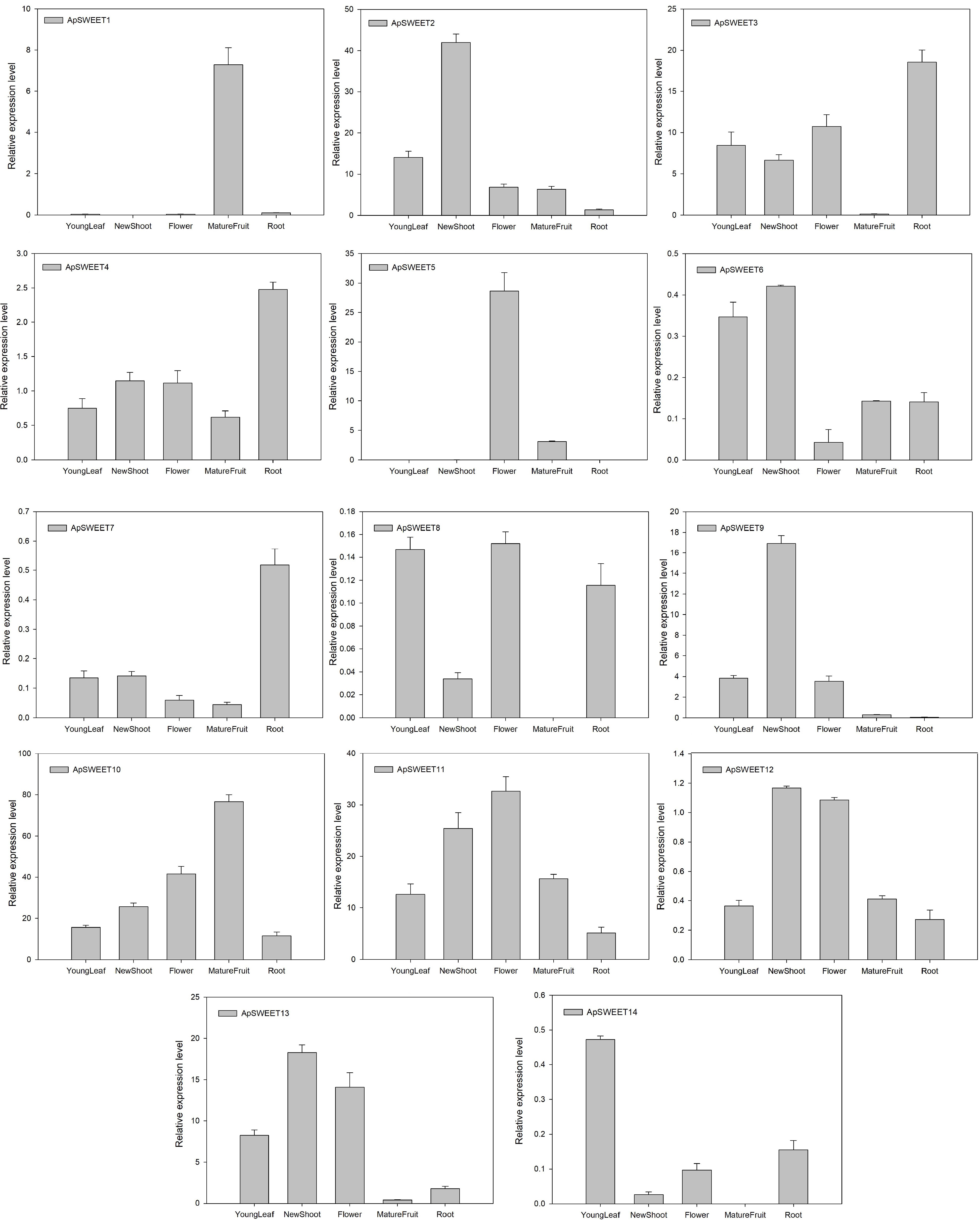
Figure 9.
ApSWEET expression of different tissues.
-
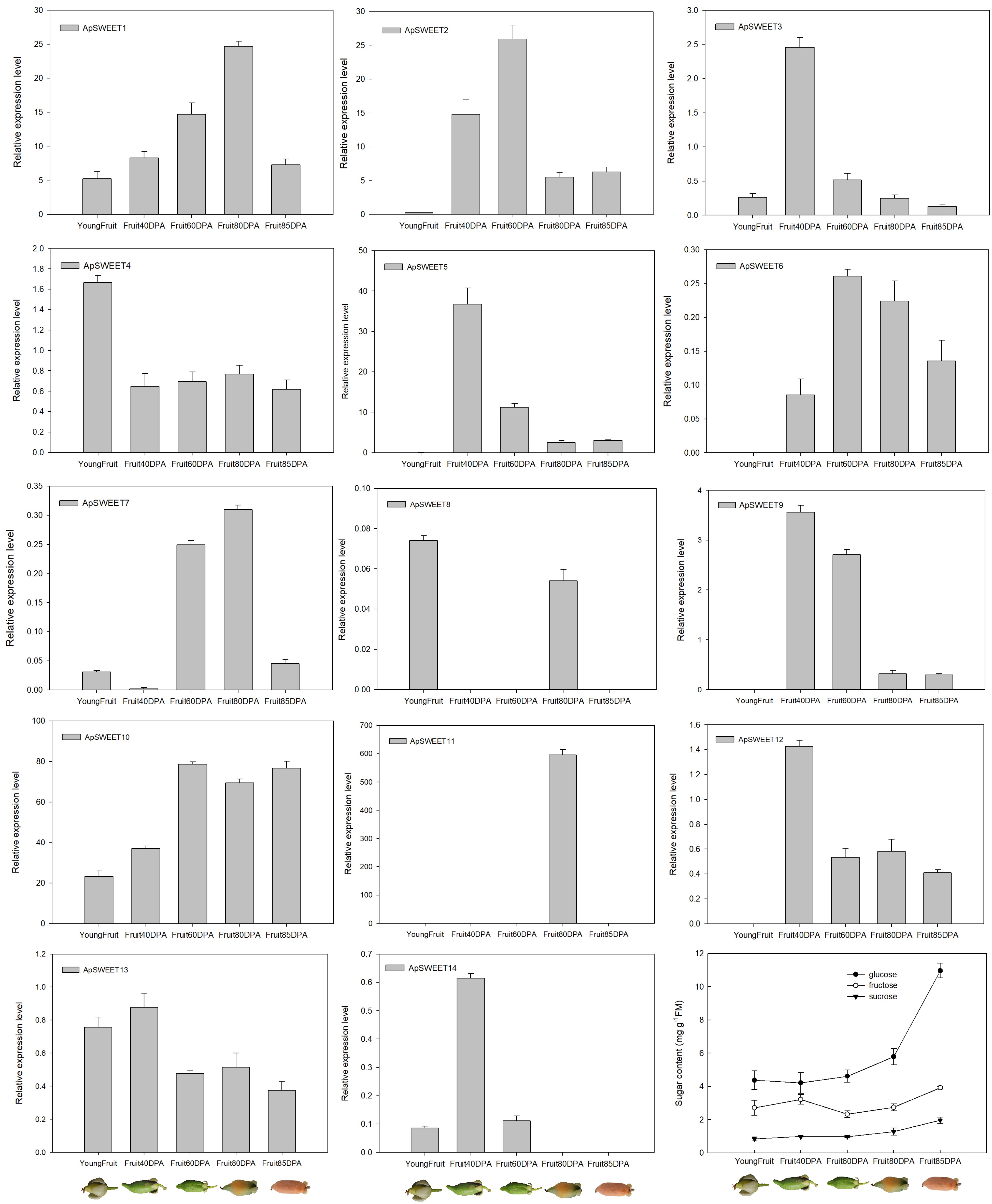
Figure 10.
ApSWEET expression of different fruit development stage.
-
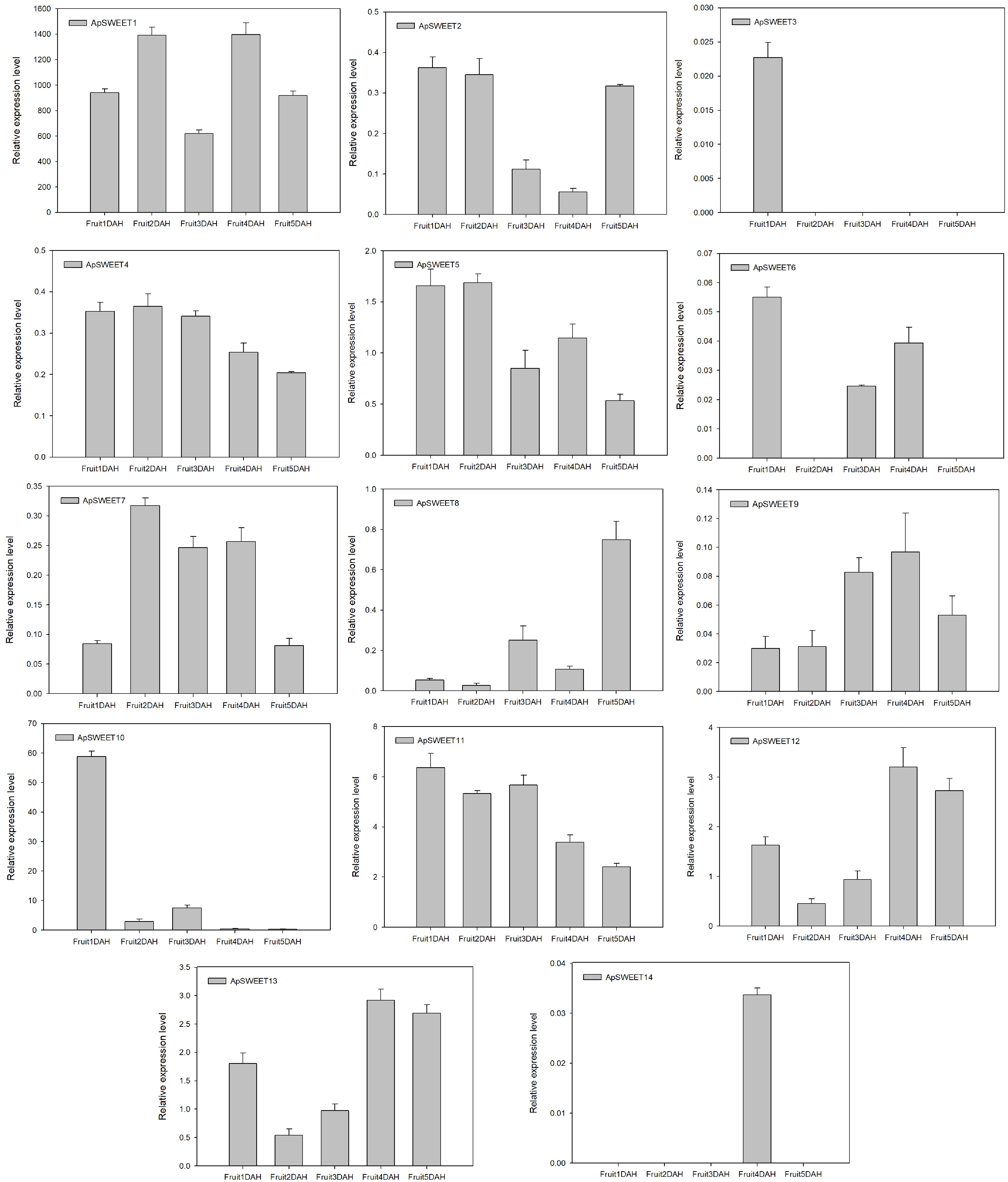
Figure 11.
ApSWEET expression of different storage periods.
-
Gene ID Number of amino acids Molecular weight pI Asp + Glu Arg + Lys Instability index Predicted location ApSWEET1 275 31,093 8.17 17 19 37.35 (stable) Cell membrane ApSWEET2 244 27,049.86 6.86 16 16 35.79 (stable) Cell membrane ApSWEET3 304 33,192.12 9.49 19 30 36.79 (stable) Chloroplast ApSWEET4 254 29,132.9 7.61 19 20 46.56 (unstable) Cell membrane ApSWEET5 259 28,602.2 9.71 13 24 45.53 (unstable) Cell membrane ApSWEET6 234 25,895.58 8.48 14 16 35.70 (stable) Cell membrane ApSWEET7 238 26,891.1 9.18 13 19 36.22 (stable) Cell membrane ApSWEET8 237 26,653.71 8.87 13 17 34.25 (stable) Cell membrane ApSWEET9 253 27,530.9 9.5 12 23 29.02 (stable) Cell membrane
Golgi apparatusApSWEET10 252 27,529.8 9.49 12 20 25.31 (stable) Cell membrane ApSWEET11 236 25,918.83 9.26 8 14 36.71 (stable) Cell membrane ApSWEET12 234 25,977.87 9.03 9 14 36.26 (stable) Cell membrane ApSWEET13 238 26,391.35 9.01 9 14 38.49 (stable) Cell membrane ApSWEET14 233 26,641.9 9.36 15 24 43.15 (unstable) Cell membrane Table 1.
Physicochemical properties of SWEET gene family proteins.
-
Alpha
helixExtended
strandBeta turn Random
coilApSWEET1 45.09% 16.73% 2.18% 36.00% ApSWEET2 41.80% 22.54% 2.87% 32.79% ApSWEET3 49.01% 15.46% 5.26% 30.26% ApSWEET4 37.80% 18.11% 0.79% 43.31% ApSWEET5 35.91% 21.24% 4.63% 38.22% ApSWEET6 50.00% 21.79% 3.85% 24.36% ApSWEET7 40.34% 21.85% 4.20% 33.61% ApSWEET8 40.51% 21.94% 5.91% 31.65% ApSWEET9 33.99% 25.30% 4.35% 36.36% ApSWEET10 46.83% 17.06% 3.97% 32.14% ApSWEET11 42.37% 19.49% 5.08% 33.05% ApSWEET12 41.88% 20.94% 4.27% 32.91% ApSWEET13 46.22% 19.33% 2.52% 31.93% ApSWEET14 36.48% 21.89% 3.86% 37.77% Table 2.
Secondary structure analysis of ApSWEET family members.
Figures
(11)
Tables
(2)