-
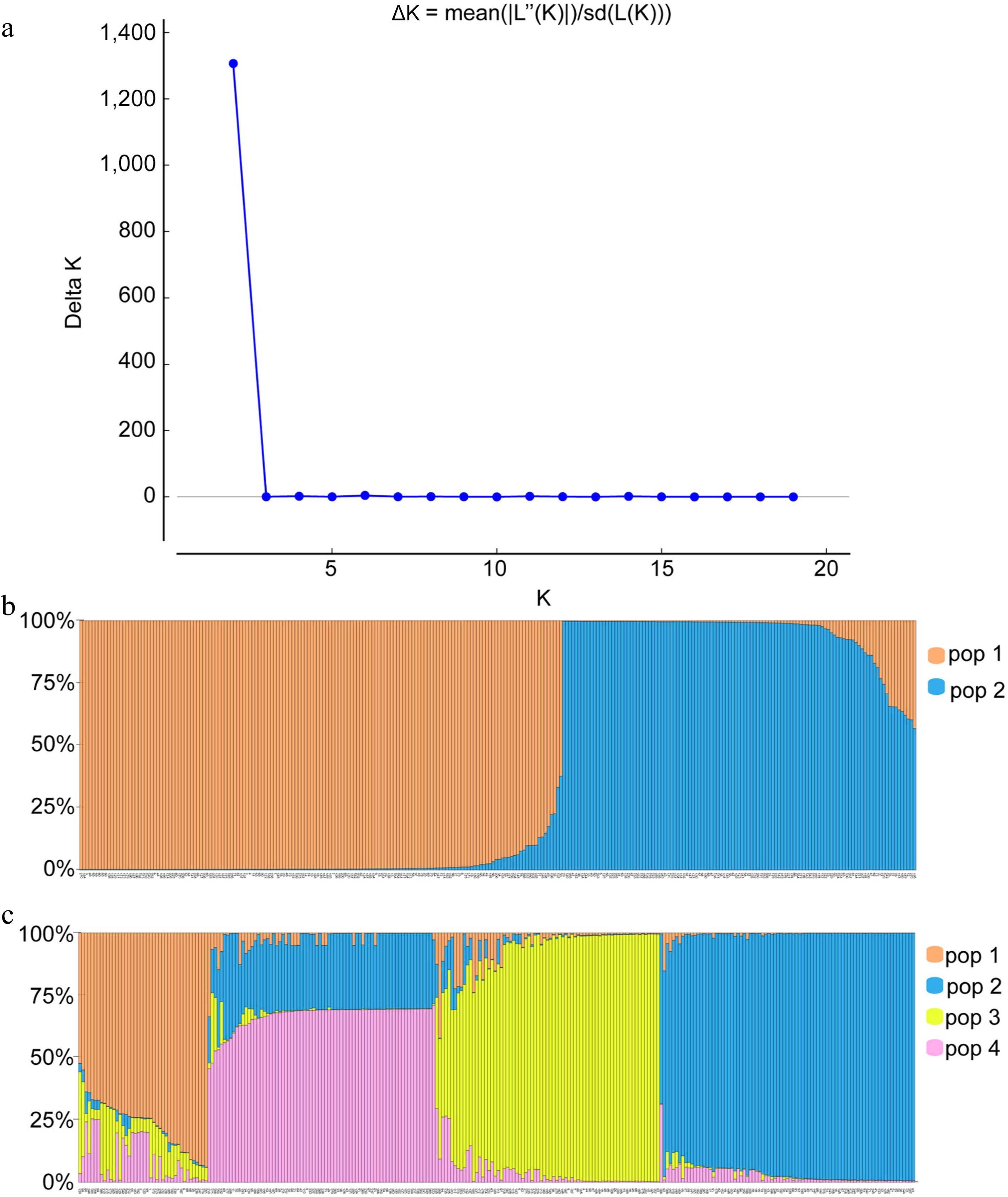
Figure 1.
Bar plots of all 272 individuals from Pueraria germplasm grouped into two or four genetic clusters with assignment probabilities obtained from STRUCTURE analyses of polymorphisms at 23 simple sequence repeat loci. (a) Distribution of delta K = 1−20. (b), (c) Histogram of the STRUCTURE assignment test when K = 2 or K = 4, respectively. The number represents the code in Supplemental Table S1.
-
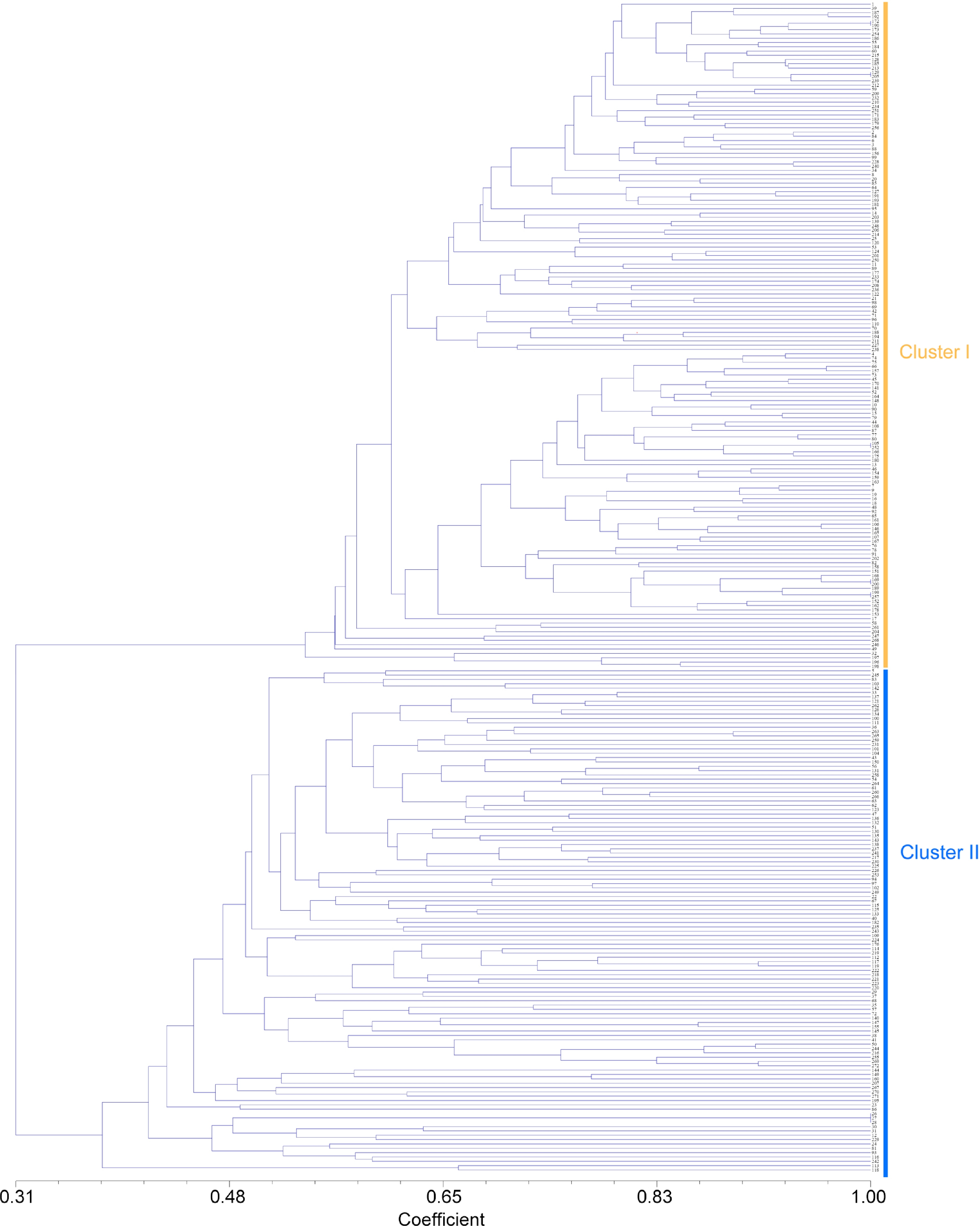
Figure 2.
Cluster diagram based on jaccard by UPGMA analysis calculated from alleles derived from 272 Pueraria accessions. The number represents the code in Supplemental Table S1.
-
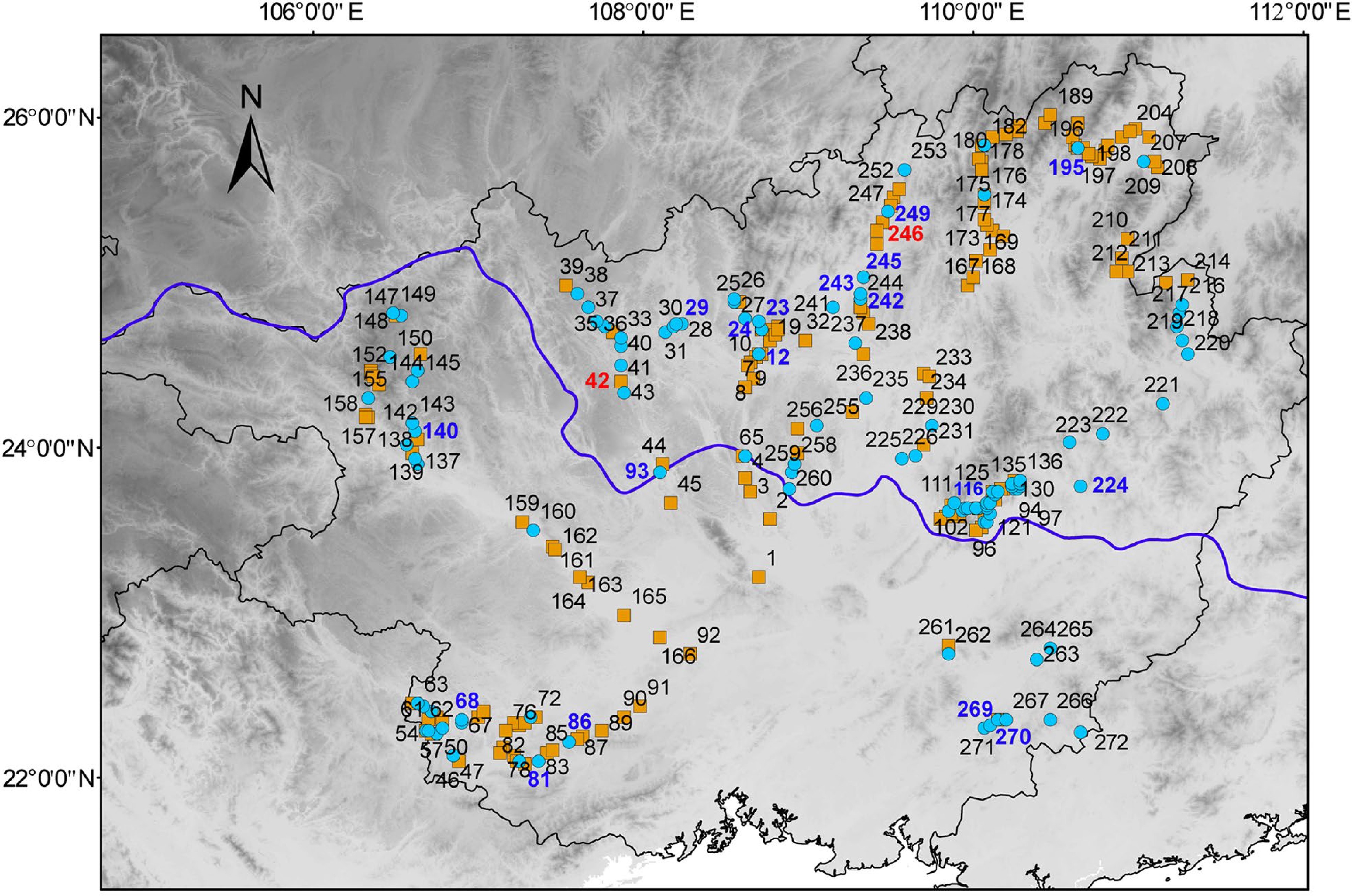
Figure 3.
Geographical distribution of the accessions collected in Guangxi. The number represents the code in Supplemental Table S1. The red and blue numbers represent two clusters of the 20 accessions of core germplasms. The orange squares represent the accessions of Cluster I and blue circles represent the accessions of Cluster II.
-
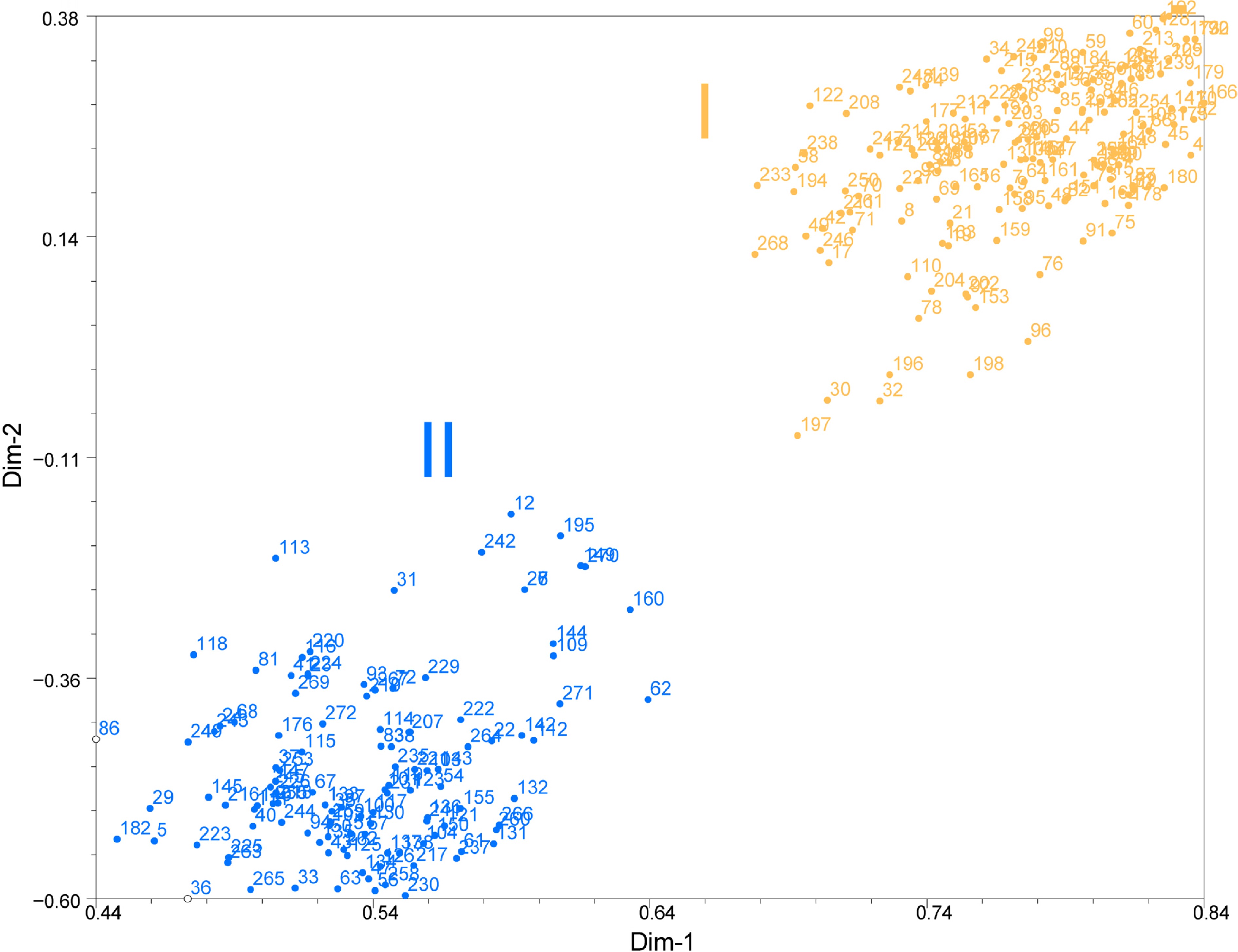
Figure 4.
PCA of Pueraria accessions based on dissimilarity matrix (Jaccard). The number represents the code in Supplemental Table S1. The number represent 272 accessions of Pueraria. The orange circles represent the accessions of Cluster I and blue circles represent the accessions of Cluster II.
-
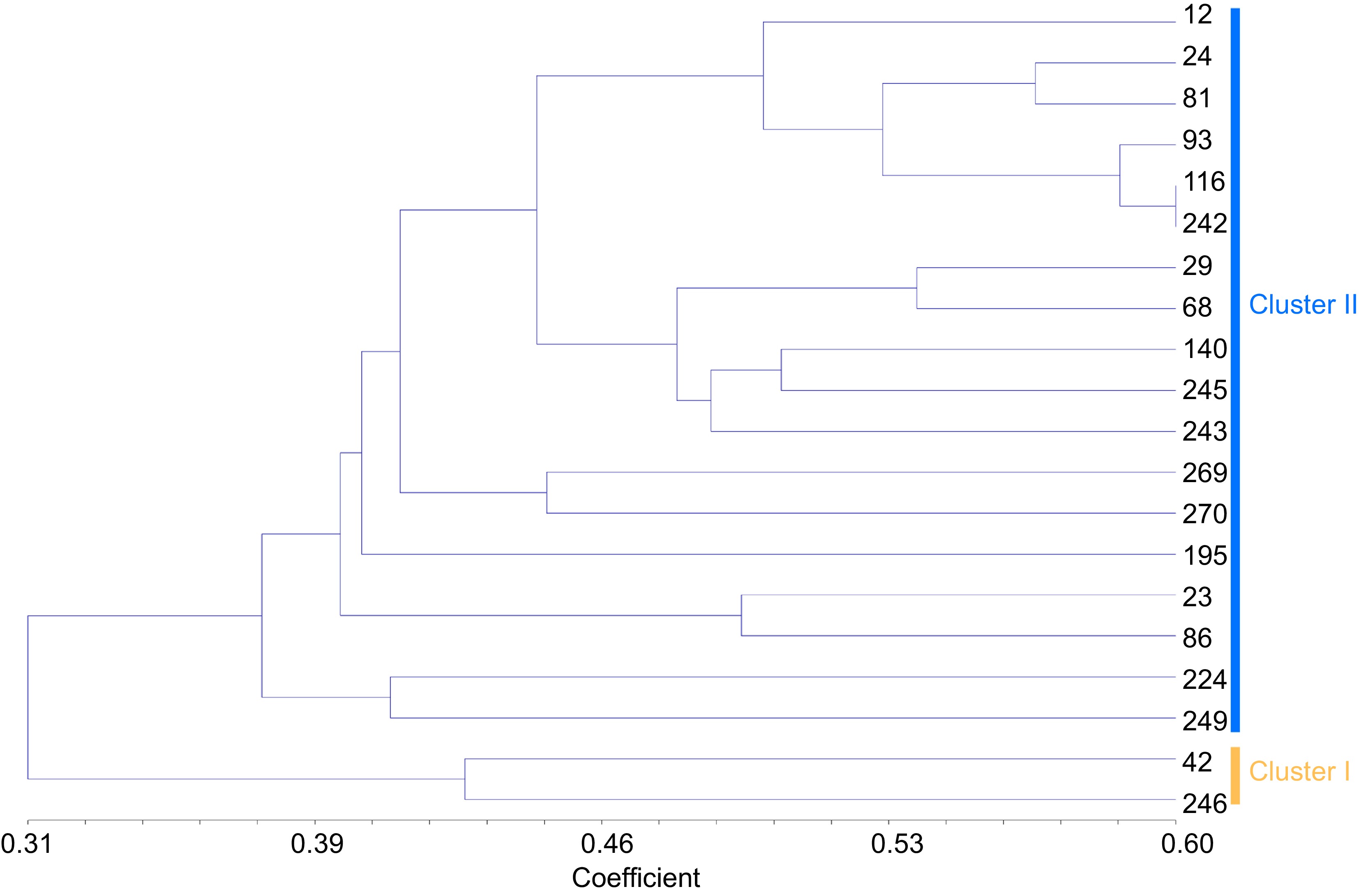
Figure 5.
Cluster diagram based on jaccard by UPGMA analysis calculated from alleles derived from 20 Pueraria accessions of core germplasm. The number represents the code in Supplemental Table S1.
-
No. Primer name Sequences (5'-3') Total number
of landsNumber of
polymorphic bandsPolymorphism
rate (%)1 PtSSR36 Fw: CTGAGTCTCTGCAAAGCCCA 10 10 100 Rv: TGTCACTGTGCTCCAACTCC 2 PtSSR98 Fw: CATTCGGACCTCCATACCCG 11 10 90.9 Rv: CCGCATCCAACCCTGATCAA 3 PtSSR99 Fw: GCTTTCCGCTGCTACCATTC 7 7 100 Rv: GCAACCCCAATGCTTCACAG 4 PtSSR104 Fw: CACCCTCCCACCACTACAAC 3 3 100 Rv: GCAATGTCCTCCTCAGCTGT 5 PtSSR108 Fw: AGCGTGCCCAACTCAGTTAA 3 3 100 Rv: CGACGGAGAAGGAGGGAATG 6 PtSSR109 Fw: CAACCTGGCTTCTGTTGTGC 5 4 80 Rv: CTCTGAAACGCTGGGCAATG 7 PtSSR121 Fw: ACACTCAACACTCCACCACC 3 2 66.67 Rv: AGGGTTTCCACCTTGAACCG 8 PtSSR122 Fw: GGGGTTTCTTCTCGGCTGAA 11 11 100 Rv: CACCCCCTTCACGCTTCATA 9 PtSSR130 Fw: ATCAGTGTCTACGTGGGGGA 5 4 80 Rv: CACTGCAGCCACAACAACAT 10 PtSSR135 Fw: GATCCGCACCCTATCTGTGG 8 8 100 Rv: CTGCGACAGCTCCGATCTTA 11 PtSSR144 Fw: TGTTGCTTTGAACACTAACATGCT 3 3 100 Rv: TGCCCTTGTCAGACACAACA 12 PtSSR155 Fw: TTCAACATTCCCCCAACCCC 2 2 100 Rv: AAGAAGAGGAACACCAGGCC 13 PtSSR168 Fw: GATCCCACCCACCACTTCTG 5 5 100 Rv: GGCTCTAGTTCTGGTGCTGG 14 PtSSR172 Fw:TCTCCAAAACAAGAAGGAAACTCC 4 3 75 Rv: TCTTTCCTCTTCTGGTATCCCA 15 PtSSR174 Fw: CAAAGAAGAAGCAGCCGCAG 6 6 100 Rv: GTCAATCCCGAAGCACTTGC 16 PtSSR175 Fw: CTGAGTCTCTGCAAAGCCCA 7 7 100 Rv: TGTCACTGTGCTCCAACTCC 17 PtSSR186 Fw: TGTTGCTTTGAACACTAACATGCT 4 4 100 Rv: TGCCCTTGTCAGACACAACA 18 PtSSR187 Fw: TGTTGCTTTGAACACTAACATGCT 4 4 100 Rv: TGCCCTTGTCAGACACAACA 19 PtSSR190 Fw: AACTGCAGGAGGAGCATGAC 5 5 100 Rv: GAGCCTCCAGGTTCTTGTCC 20 PtSSR191 Fw: GGAAGCATTGCGGTTTGGTT 3 3 100 Rv: TCACATCACATGCTGCCACT 21 PtSSR196 Fw: GCAAGAACCTGTGCTCCTCT 3 2 66.67 Rv: TGCCAATGCCATTGTGGTTG 22 PtSSR201 Fw: GCCTCTTCCAGCGAGAACTT 4 4 100 Rv: TGATCCTCCCCAACAAGCTG 23 PtSSR222 Fw: TGTGCAAGAAGGATGGGTGA 2 2 100 Rv: GGTTGCATTCGGAAGCAACA Total 118 112 Avarage 5.13 4.87 94.91 Table 1.
Amplification results and polymorphism information of 23 SSR primers.
-
Locus Sample size Na Ne h I Ht Hs Gst Nm 36-1 272 2 1.0112 0.011 0.0344 0.0129 0.0128 0.0066 75.8301 36-2 272 2 1.0112 0.011 0.0344 0.0129 0.0128 0.0066 75.8301 36-3 272 2 1.0463 0.0443 0.1082 0.0517 0.0503 0.0273 17.8197 36-4 272 2 1.0074 0.0074 0.0244 0.0086 0.0086 0.0043 114.4978 36-5 272 2 1.1017 0.0923 0.1941 0.1063 0.1007 0.0531 8.9206 36-6 272 2 1.0843 0.0778 0.1696 0.0905 0.086 0.0499 9.5227 36-7 272 2 1.2654 0.2098 0.3650 0.2394 0.2025 0.1541 2.7446 36-8 272 2 1.9322 0.4824 0.6755 0.4915 0.4157 0.1543 2.7395 36-9 272 2 1.034 0.0329 0.0849 0.0309 0.0308 0.0032 156.1603 36-10 272 2 1.0074 0.0074 0.0244 0.0086 0.0086 0.0043 114.4978 98-1 272 2 1.0188 0.0184 0.0527 0.0216 0.0213 0.011 44.8945 98-2 272 2 1.1442 0.126 0.2473 0.145 0.1336 0.0786 5.8651 98-3 272 2 1.1499 0.1304 0.2539 0.1509 0.1374 0.0895 5.0838 98-4 272 2 1.0584 0.0552 0.1291 0.0634 0.0616 0.0275 17.6531 98-5 272 2 1.0383 0.0369 0.0933 0.0431 0.0421 0.0225 21.6887 98-6 272 2 1.0383 0.0369 0.0933 0.0431 0.0421 0.0225 21.6887 98-7 272 2 1.0671 0.0629 0.1433 0.0733 0.0704 0.0396 12.1273 98-8 272 2 1.0671 0.0629 0.1433 0.0733 0.0704 0.0396 12.1273 98-9 272 2 1.16 0.1379 0.2652 0.1595 0.1442 0.0957 4.7224 98-10 272 1 1.0000 0.0000 0.0000 0 0 98-11 272 2 1.0149 0.0147 0.0436 0.0161 0.016 0.003 166.8934 99-1 272 2 1.0188 0.0184 0.0527 0.0216 0.0213 0.011 44.8945 99-2 272 2 1.0615 0.058 0.1343 0.0592 0.0592 0.0007 711.3620 99-3 272 2 1.288 0.2236 0.3830 0.2029 0.1857 0.0849 5.3899 99-4 272 2 1.2065 0.1712 0.3129 0.1855 0.1781 0.04 12.0065 99-5 272 2 1.14 0.1228 0.2424 0.1422 0.1304 0.0835 5.4886 99-6 272 2 1.3643 0.267 0.4375 0.2622 0.261 0.0043 114.6222 99-7 272 2 1.5399 0.3506 0.5353 0.3448 0.3421 0.0076 65.5416 104-1 272 2 1.034 0.0329 0.0850 0.0352 0.0351 0.0039 128.2046 104-2 272 2 1.9558 0.4887 0.6818 0.4964 0.3986 0.1969 2.0392 104-3 272 2 1.9992 0.4998 0.6929 0.4957 0.3738 0.2459 1.5335 108-1 272 2 1.6292 0.3862 0.5746 0.393 0.3882 0.0122 40.3512 108-2 272 2 1.4728 0.321 0.5016 0.3331 0.3234 0.0292 16.6414 108-3 272 2 1.6831 0.4058 0.5958 0.3972 0.3883 0.0223 21.9303 109-1 272 2 1.2934 0.2269 0.3872 0.2568 0.2167 0.1562 2.7010 109-2 272 2 1.0149 0.0147 0.0436 0.0161 0.016 0.003 166.8934 109-3 272 2 1.2934 0.2269 0.3872 0.2568 0.2167 0.1562 2.7010 109-4 272 2 1.0149 0.0147 0.0436 0.0161 0.016 0.003 166.8934 109-5 272 1 1 0 0.0000 0 0 121-1 272 2 1.0753 0.07 0.1561 0.0805 0.0775 0.0374 12.8581 121-2 272 2 1.0753 0.07 0.1561 0.0805 0.0775 0.0374 12.8581 121-3 272 1 1 0 0.0000 0 0 122-1 272 2 1.0304 0.0295 0.0778 0.0345 0.0339 0.0179 27.4911 122-2 272 2 1.1351 0.119 0.2367 0.1379 0.1268 0.0805 5.7097 122-3 272 2 1.0304 0.0295 0.0778 0.0345 0.0339 0.0179 27.4911 122-4 272 2 1.0887 0.0815 0.1760 0.0948 0.0898 0.0525 9.0192 122-5 272 2 1.0887 0.0815 0.1760 0.0948 0.0898 0.0525 9.0192 122-6 272 2 1.0671 0.0629 0.1433 0.0733 0.0704 0.0396 12.1273 122-7 272 2 1.0343 0.0332 0.0856 0.0388 0.038 0.0202 24.2677 122-8 272 2 1.4253 0.2984 0.4752 0.2929 0.2912 0.0058 85.4766 122-9 272 2 1.0037 0.0037 0.0134 0.0043 0.0043 0.0022 230.4989 122-10 272 2 1.5187 0.3415 0.5250 0.3673 0.3146 0.1435 2.9847 122-11 272 2 1.2973 0.2291 0.3902 0.2608 0.2152 0.1748 2.3604 130-1 272 2 1.2025 0.1684 0.3090 0.194 0.1703 0.1221 3.5945 130-2 272 2 1.2025 0.1684 0.3090 0.194 0.1703 0.1221 3.5945 130-3 272 2 1.0383 0.0369 0.0933 0.0431 0.0421 0.0225 21.6887 130-4 272 2 1.0383 0.0369 0.0933 0.0431 0.0421 0.0225 21.6887 130-5 272 1 1 0 0.0000 0 0 135-1 272 2 1.0149 0.0147 0.0438 0.0172 0.0171 0.0088 56.4956 135-2 272 2 1.0587 0.0554 0.1296 0.0647 0.0624 0.0346 13.9494 135-3 272 2 1.3477 0.258 0.4264 0.2931 0.2295 0.2171 1.8032 135-4 272 2 1.0111 0.011 0.0343 0.0118 0.0118 0.0013 397.0141 135-5 272 2 1.3127 0.2382 0.4017 0.2716 0.219 0.1934 2.0859 135-6 272 2 1.0149 0.0147 0.0436 0.0161 0.016 0.003 166.8934 135-7 272 2 1.7785 0.4377 0.6295 0.4706 0.1885 0.5994 0.3341 135-8 272 2 1.9656 0.4913 0.6844 0.4999 0.1558 0.6884 0.2263 144-1 272 2 1.0343 0.0332 0.0856 0.0388 0.038 0.0202 24.2677 144-2 272 2 1.9251 0.4805 0.6736 0.479 0.4776 0.0029 174.8282 144-3 272 2 1.6998 0.4117 0.6020 0.3877 0.3213 0.1713 2.4193 155-1 272 2 1.0698 0.0652 0.1475 0.0687 0.0684 0.0049 101.9746 155-2 272 2 1.2483 0.1989 0.3507 0.2281 0.1936 0.1512 2.8079 168-1 272 2 1.501 0.3338 0.5162 0.3124 0.2825 0.0956 4.7296 168-2 272 2 1.9254 0.4806 0.6736 0.4892 0.4321 0.1166 3.7876 168-3 272 2 1.5113 0.3383 0.5214 0.3181 0.2904 0.0869 5.2557 168-4 272 2 1.9569 0.489 0.6821 0.4954 0.4328 0.1262 3.4613 168-5 272 2 1.0074 0.0073 0.0243 0.0075 0.0075 0.0001 2000.0000 172-1 272 2 1.2368 0.1915 0.3407 0.2198 0.1882 0.1438 2.9771 172-2 272 2 1.0112 0.011 0.0344 0.0129 0.0128 0.0066 75.8301 172-3 272 1 1 0 0.0000 0 0 172-4 272 2 1.0037 0.0037 0.0134 0.0032 0.0032 0.0016 310.4992 174-1 272 2 1.0074 0.0074 0.0244 0.0086 0.0086 0.0043 114.4978 174-2 272 2 1.074 0.0689 0.1542 0.0741 0.0734 0.0099 50.0991 174-3 272 2 1.1249 0.111 0.2241 0.1278 0.1192 0.0669 6.9725 174-4 272 2 1.0037 0.0037 0.0134 0.0043 0.0043 0.0022 230.4989 174-5 272 2 1.7565 0.4307 0.6221 0.4621 0.2446 0.4706 0.5625 174-6 272 2 1.9722 0.4929 0.6861 0.5 0.2054 0.5892 0.3486 175-1 272 2 1.0074 0.0074 0.0244 0.0086 0.0086 0.0043 114.4978 175-2 272 2 1.0932 0.0852 0.1823 0.0991 0.0937 0.0552 8.5594 175-3 272 2 1.0074 0.0074 0.0244 0.0086 0.0086 0.0043 114.4978 175-4 272 2 1.2069 0.1714 0.3132 0.1965 0.1735 0.117 3.7725 175-5 272 2 1.8504 0.4596 0.6522 0.4769 0.3654 0.2337 1.6394 175-6 272 2 1.7047 0.4134 0.6038 0.39 0.3252 0.1659 2.5132 175-7 272 2 1.0301 0.0292 0.0771 0.0298 0.0298 0.0003 1480.2742 186-1 272 2 1.0421 0.0404 0.1005 0.0462 0.0453 0.0181 27.1466 186-2 272 2 1.7726 0.4359 0.6276 0.4059 0.2739 0.3252 1.0373 186-3 272 2 1.7848 0.4397 0.6316 0.4698 0.2325 0.5051 0.4899 186-4 272 2 1.6977 0.411 0.6013 0.3808 0.2793 0.2664 1.3765 187-1 272 2 1.042 0.0403 0.1002 0.0449 0.0444 0.0122 40.4142 187-2 272 2 1.7641 0.4331 0.6247 0.4021 0.2655 0.3397 0.9718 187-3 272 2 1.875 0.4667 0.6594 0.4905 0.1571 0.6796 0.2357 187-4 272 2 1.6636 0.3989 0.5883 0.3687 0.2779 0.2465 1.5286 190-1 272 2 1.0993 0.0903 0.1908 0.0833 0.082 0.0164 29.9125 190-2 272 2 1.046 0.044 0.1076 0.0385 0.0377 0.02 24.4900 190-3 272 2 1.1535 0.1331 0.2580 0.1211 0.1166 0.037 13.0124 190-4 272 2 1.6024 0.3759 0.5634 0.4076 0.2996 0.2651 1.3862 190-5 272 2 1.9999 0.5 0.6931 0.4956 0.323 0.3483 0.9354 191-1 272 2 1.9284 0.4814 0.6745 0.498 0.1126 0.7738 0.1462 191-2 272 2 1.6789 0.4044 0.5942 0.3708 0.2541 0.3146 1.0893 191-3 272 2 1.1641 0.141 0.2696 0.1261 0.1192 0.0552 8.5656 196-1 272 2 1.2295 0.1867 0.3342 0.2136 0.1856 0.1312 3.3100 196-2 272 2 1.0074 0.0074 0.0244 0.0086 0.0086 0.0043 114.4978 196-3 272 1 1 0 0.0000 0 0 201-1 272 2 1.0037 0.0037 0.0134 0.0043 0.0043 0.0022 230.4989 201-2 272 2 1.9978 0.4995 0.6926 0.4989 0.3531 0.2923 1.2103 201-3 272 2 1.404 0.2877 0.4625 0.3224 0.2509 0.2219 1.7536 201-4 272 2 1.4033 0.2874 0.4621 0.2798 0.2768 0.011 45.0326 222-1 272 2 1.3039 0.233 0.3951 0.2651 0.2176 0.1791 2.2910 222-2 272 2 1.7711 0.4354 0.6271 0.4617 0.3031 0.3435 0.9556 Mean 272 1.9492 1.2841 0.1778 0.2858 0.1841 0.1435 0.2204 1.7690 St. Dev 0.2206 0.3277 0.1741 0.2397 0.0305 0.0166 Na = Observed number of alleles; Ne = Effective number of alleles[56]; h = Nei's (1973) gene diversity; I = Shannon's Information index[57]; Gst = coefficient of gene differentiation; Nm = estimate of gene flow from Gst or Gcs. E.g., Nm = 0.5(1 - Gst)/Gst; Ht = Total expected heterozygosity; Hs = the average expected heterozygosity within subpopulations. Table 2.
Genetic characteristics for 112 polymorphic microsatellite loci in 272 individuals of Pueraria species in the present study.
-
Sampling proportion Sample number Na Ne h I Number of polymorphic loci Percentage of polymorphic loci Percentage of total loci 5% 14 1.8644 ± 0.3438 1.3839 ± 0.3116 0.2413 ± 0.1639 0.3778 ± 0.2243 102 91.07% 86.44% 7.00% 19 1.8644 ± 0.3438 1.3779 ± 0.3123 0.2381 ± 0.1635 0.3736 ± 0.2242 102 91.07% 86.44% 7.35% 20 1.8898 ± 0.3144 1.3716 ± 0.3084 0.2359 ± 0.1598 0.3727 ± 0.2170 105 93.75% 88.98% 7.70% 21 1.8983 ± 0.3035 1.3658 ± 0.3063 0.2333 ± 0.1580 0.3702 ± 0.2139 106 94.64% 89.83% 8% 22 1.8983 ± 0.3035 1.3655 ± 0.3049 0.2333 ± 0.1581 0.3699 ± 0.2145 106 94.64% 89.83% 10% 27 1.8983 ± 0.3035 1.3577 ± 0.2991 0.2297 ± 0.1575 0.3648 ± 0.2154 106 94.64% 89.83% 15% 41 1.9068 ± 0.2920 1.3451 ± 0.3055 0.2208 ± 0.1613 0.3518 ± 0.2203 107 95.54% 90.68% 20% 54 1.9237 ± 0.2666 1.3384 ± 0.3106 0.2161 ± 0.1621 0.3459 ± 0.2195 109 97.32% 92.37% 30% 82 1.9322 ± 0.2525 1.3204 ± 0.3052 0.2060 ± 0.1625 0.3314 ± 0.2217 110 98.21% 93.22% 40% 109 1.9407 ± 0.2372 1.3180 ± 0.3158 0.2024 ± 0.1664 0.3249 ± 0.2272 111 99.11% 94.07% 50% 136 1.9322 ± 0.2525 1.3146 ± 0.3273 0.1980 ± 0.1703 0.3171 ± 0.2327 110 98.21% 93.22% 100% 272 1.9492 ± 0.2206 1.2841 ± 0.3277 0.1778 ± 0.1741 0.2858 ± 0.2397 112 94.92% Na = Observed number of alleles; Ne = Effective number of alleles[56]; h = Nei's (1973) gene diversity; I = Shannon's Information index[57]. Table 3.
Summary of the extraction of a core collection.
Figures
(5)
Tables
(3)