-
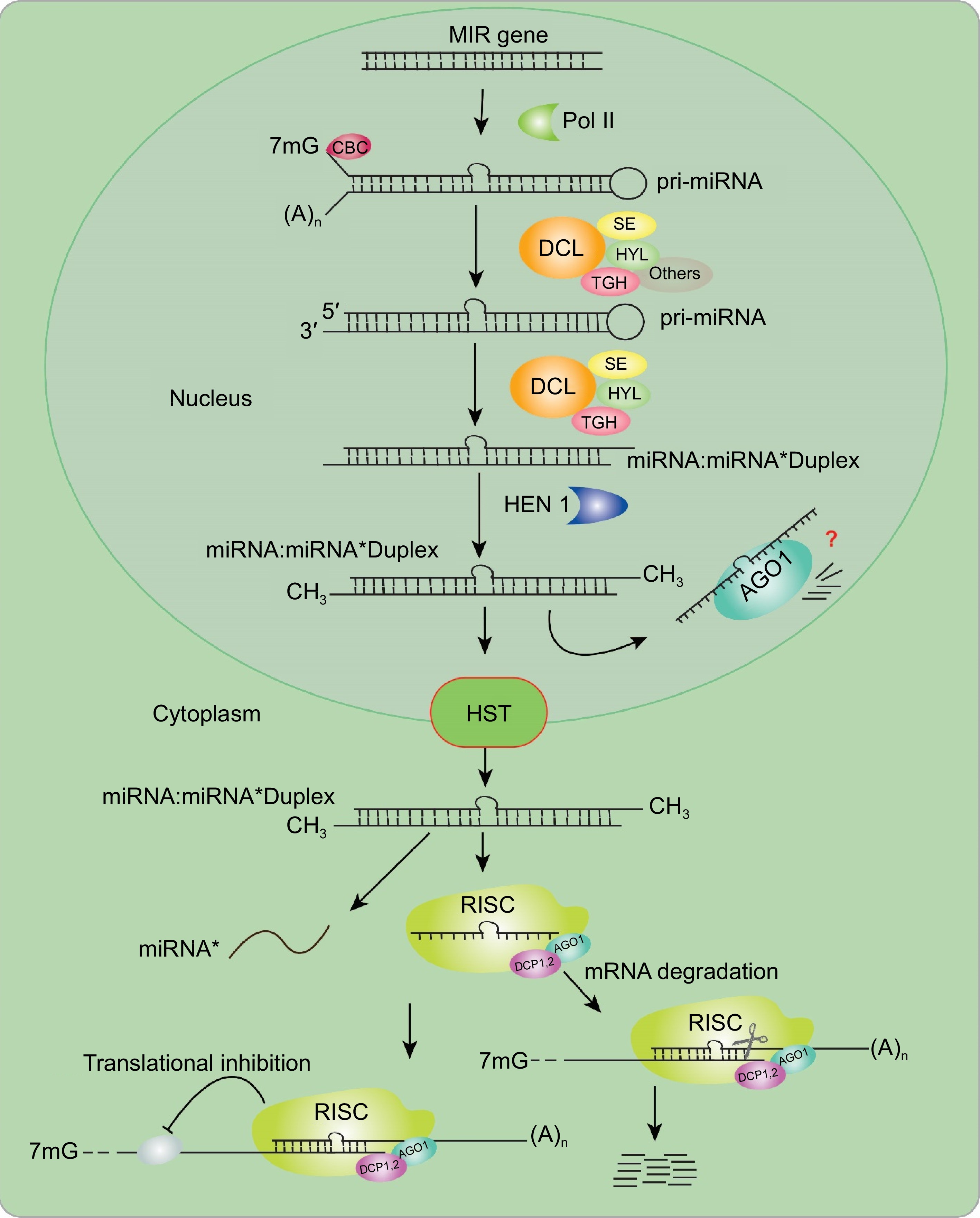
Figure 1.
Biogenesis of model miRNAs in plants. MIR genes are initially transcribed by RNA Pol II into pri-miRNAs, which then adopt hairpin structures through self-folding. The subsequent processing and splicing of miRNAs require the interaction of DDL, HYL1, SE, and TGH in conjunction with the cap-binding complex (CBC). DCL1 processes both pri-miRNAs and pre-miRNAs, generating one or several miRNA/miRNA* duplexes, subsequently methylated by HEN1. These processed miRNAs are transported to the cytoplasm by HST1. Subsequent processing involves the integration of the selected miRNAs into the RISC, which comprises AGO1. This complex guide either the inhibition of translation or the cleavage of the target mRNA transcript. AGO, Argonaute; DCL, Dicer-like; HYL1, HYPONASTIC LEAVES1; Pol II, RNA polymerase II; HEN1, HUA ENHANCER 1; CBC, cap-binding complex; MIR, MIRNA; RISC, RNA-induced silencing complex; miRNA, microRNA; mRNA, messenger RNA; SE, SERRATE.
-
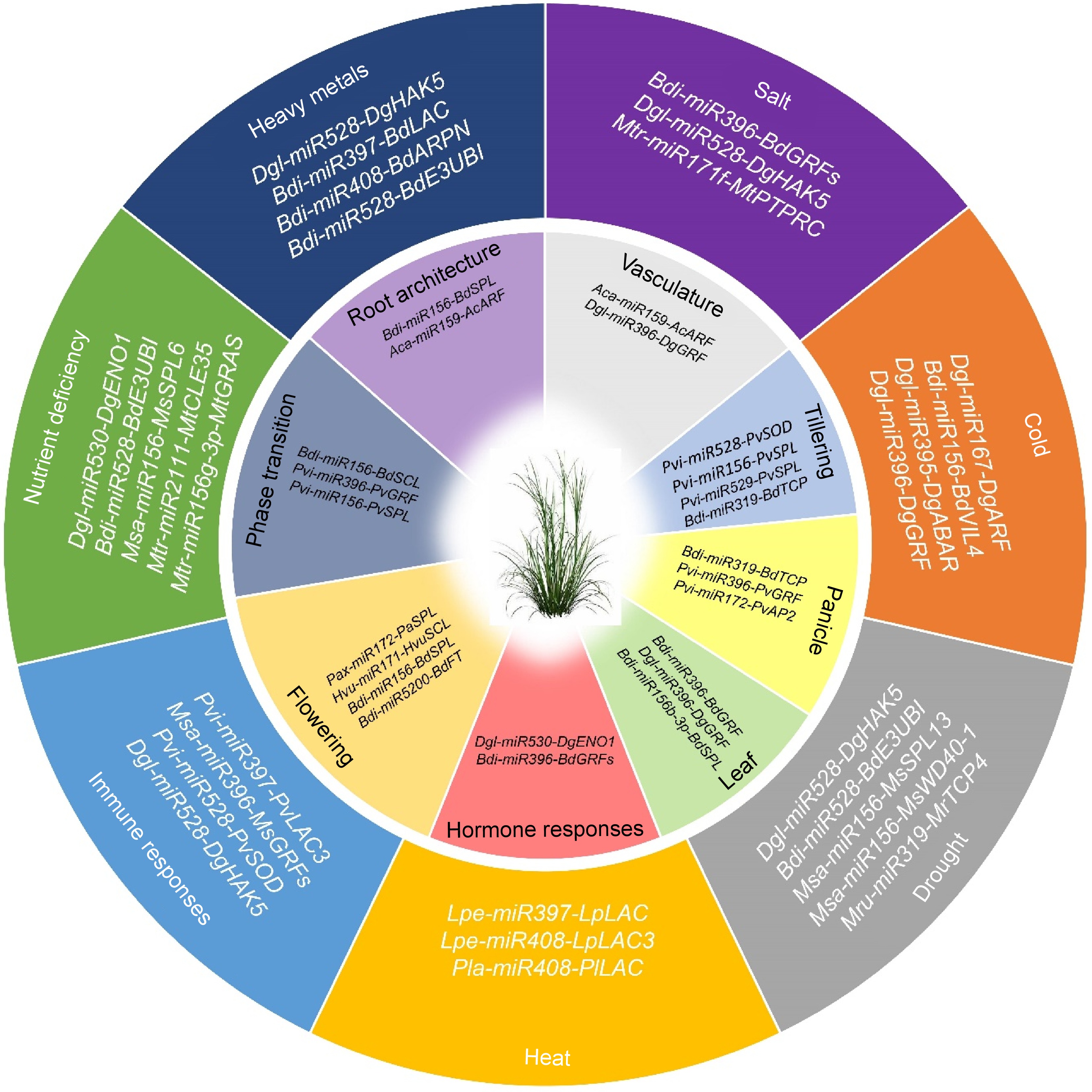
Figure 2.
MicroRNAs play pivotal role in development and stress responses in grasses. Recent understanding of miRNA-mediated regulation is outlined, encompassing development (inner circle) and responses to biotic and abiotic stresses (outer circle) in grasses. Bdi, Brachypodium distachyon; Msa, Medicago sativa; Pvi, Panicum virgatum; Pla, Paeonia lactiflora; Agr, Apium graveolens; Lpe, Lolium perenne; Hvu, Hordeum vulgare; Aca, Agrostis canina; Cda, Cynodon dactylon; Dgl, Dactylis glomerata; Mru, Medicago ruthenica, and Mtr, Medicago truncatula.
-
miRNA Species Target mRNA Biological function Ref. Bdi-miR156 Brachypodium distachyon BdSPL Root development/Leaf development/Flowering [25] Aca-miR159 Agrostis canina AcARF Root development/Vasculature [52] Dgi-miR396 Dactylis glomerata DgGRF Vasculature/Leaf development [63] Pvi-miR528 Panicum virgatum PvSOD Tillering [33] Pvi-miR156 Panicum virgatum PvSPL Tillering [24,25] Pvi-miR529 Panicum virgatum PvSPL Tillering [24] Bdi-miR319 Brachypodium distachyon BdTCP Tillering/Panicle [61] Pvi-miR396 Panicum virgatum PvGRF Panicle [37] Pvi-miR172 Panicum virgatum PvAP2 Panicle [24,25,27] Bdi-miR396 Brachypodium distachyon BdGRF Leaf development/Hormone response [37] Bdi-miR5200 Brachypodium distachyon BdFT Flowering [35] Hvu-miR171 Hordeum vulgare HvSCL Flowering [26] Pax-miR172 petunia PaSPL Flowering [27] Table 1.
List of miRNAs involved in grasses development.
-
miRNA Species Target mRNA Biological function Ref. Bdi-miR396 Brachypodium distachyon BdGRF Salt [37] Dgl-miR528 Dactylis glomerata DgHAK5 Salt/Drought/Immune response/Heavy metals [33] Mtr-miR171f Medicago truncatula MtPTPRC Salt [52] Dgl-miR167 Dactylis glomerata DgARF Cold [63] Bdi-miR156 Brachypodium distachyon BdVIL4 Cold [62] Dgl-miR395 Dactylis glomerata DgABAR Cold [58] Dgl-miR396 Dactylis glomerata DgGRF Cold [37] Bdi-miR528 Brachypodium distachyon BdE3UBI Drought/Nutrient deficiency/Heavy metals [56] Msa-miR156 Medicago sativa MsSPL13 Drought [60] Msa-miR156 Medicago sativa MsWD40-1 Drought [60] Mru-miR319 Medicago ruthenica MrTCP4 Drought [61] Lpe-miR397 Lolium perenne LpLAC Heat [51] Lpe-miR408 Lolium perenne LpLAC3 Heat [50] Pla-miR408 Paeonia lactiflora PlLAC Heat [47] Pvi-miR397 Panicum virgatum PvLAC Immune response [51] Msa-miR396 Medicago sativa MsGRF Immune response [43] Pvi-miR528 Panicum virgatum PvSOD Immune response [33] Dgl-miR530 Dactylis glomerata DgENO1 Nutrient deficiency [63] Msa-miR156 Medicago sativa MsSPL Nutrient deficiency [44] Mtr-miR2111 Medicago truncatula MtCLE35 Nutrient deficiency [14] Mtr-miR156g-3p Medicago truncatula MtGRAS Nutrient deficiency [45] Bdi-miR397 Brachypodium distachyon BdLAC Heavy metals [51] Bdi-miR408 Brachypodium distachyon BdARPN Heavy metals [51,59] Table 2.
List of miRNAs involved in responses to biotic and abiotic stresses.
Figures
(2)
Tables
(2)