-
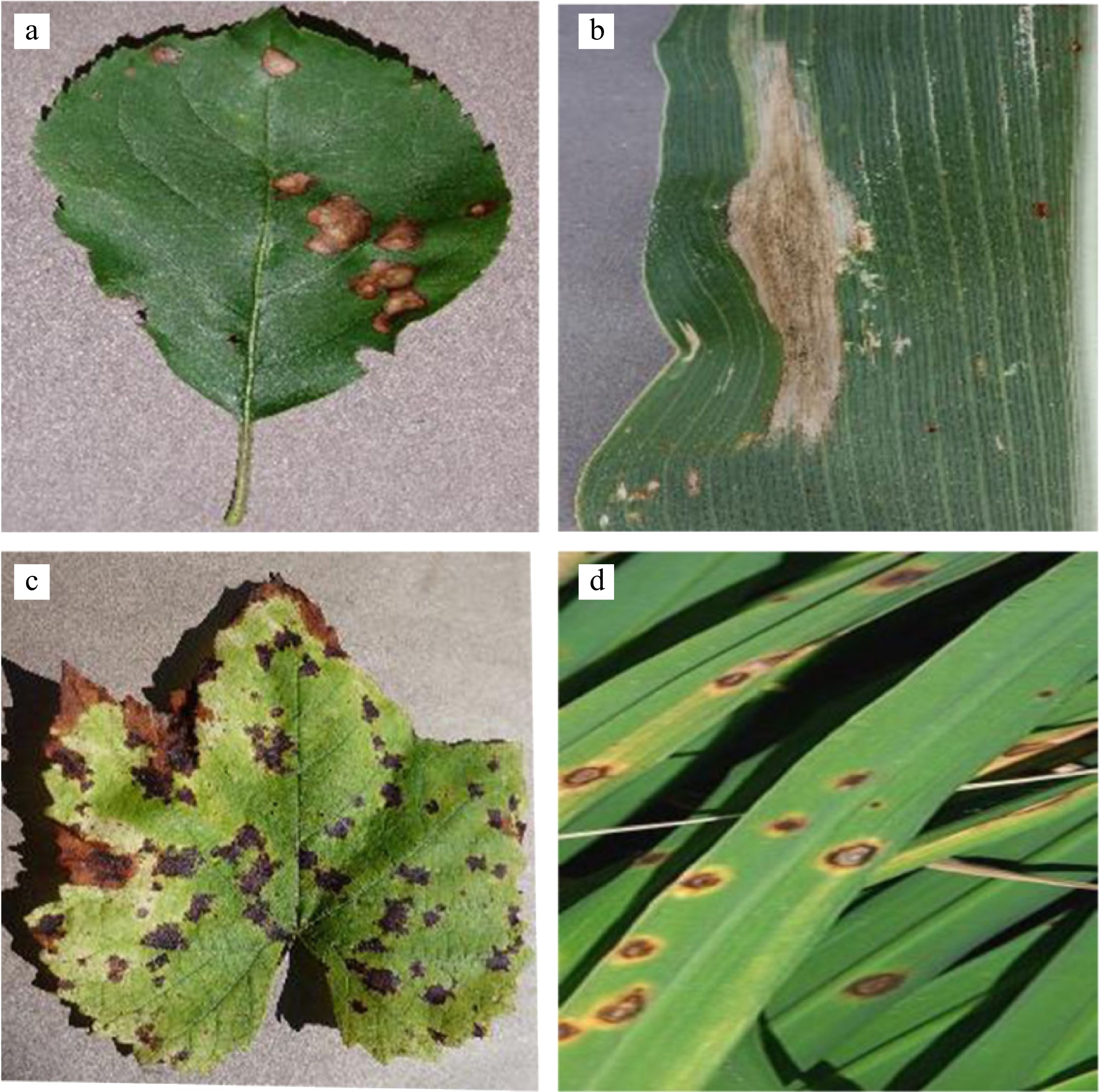
Figure 1.
Photographic examples of a plant disease dataset. (a) Balck rot; (b) Northern leaf blight; (c) Isariopsis leaf spot; (d) Brown spot.
-
Data set name Crop Brief description Ref. Image Database of Plant Disease Symptoms 21 plant species In October of 2016, this database called PDDB, had 2,326 images of 171 diseases and other disorders affecting 21 plant species including soybean, citrus, coconut tree, dry bean, cassava, passion fruit, corn, coffee, cashew tree, grapevine, oil palm, wheat, sugarcane, cotton, black pepper, cabbage, melon, rice, pineapple, papaya, cupuacu. [26] Tomato leaf disease detection Tomato A tomato leaf disease dataset includes tomato mosaic virus, tomato yellow leaf curl virus, late blight, leaf mold, early blight, septoria leaf spot, Spider mites Two-spotted spider mite, healthy tomato. [27] Agricultural Disease Image Database for Agricultural Diseases and Pests Research Rice and wheat, fruits
and vegetables, etc.The dataset currently has about 15,000 high-quality agricultural disease images, including field crops such as rice and wheat, fruits and vegetables such as cucumber and grape, etc. [28] Rice disease dataset Rice A rice leaf disease dataset includes bacterial leaf blight, blast and brown spot. [29] Pathological images of apple leaves Apple The apple leaf disease image dataset contains 8 common apple leaf diseases: Mosaic, rust, gray spot, mottle leaf disease, brown spot, black star disease, black rot, healthy leaf disease. [30] Plant Pathology 2021 - FGVC8 Apple 2021-FGVC8 contains approximately 23,000 high-quality RGB images of apple foliar diseases, including a large expert-annotated disease dataset. [31] Tomato Disease Multiple Sources Tomato Over 20 k images of tomato leaves with 10 diseases and one healthy class. Images are collected from both lab scenes and in-the-wild scenes, which includes late blight, healthy, early blight, septoria leaf spot, tomato yellow leaf curl virus, bacterial spot, target spot, tomato mosaic virus, leaf mold, spider mites Two-spotted spider mite, powdery mildew. [32] Data for: Identification of Plant Leaf Diseases Using a 9-layer Deep Convolutional Neural Network 12 plant species The dataset includes 39 different classes of plant leaf and background images are available. The data-set contain 61,486 images. [33] Table 1.
Plant disease open data sets.
-
No. Model name Technical characteristics Advantages. Ref. 1 Apple-Net The Feature Enhancement Module (FEM) and Coordinate Attention (CA) incorporation.
Generative Adversarial Networks (GAN) for interference reduction.Multi-scale information acquisition, increased diversity, and noise resistance. [34] 2 Mobile Ghost with Attention YOLO Ghost modules and separable convolution for reducing model size.
The mobile inverted residual bottleneck convolution with Convolutional Block Attention Module (CBAM) for improving feature extraction capability.Lightweight real-time monitoring (10.34 MB), suitable for mobile terminals. [35] 3 BTC-YOLOv5s Bidirectional Feature Pyramid Network (BiFPN) for a fusion of multi-scale features.
Transformer attention mechanism for capturing global contextual information and establishing long-range dependencies.
CBAM for interference reduction.Reduces irrelevant information, small model size (15.8 MB). [36] 4 AlAD-YOLO The backbone network of TOLOv5s replaced with that of MobileNetV3. Reduction in parameters and computational complexity during feature extraction. [37] 5 YOLOX-ASSANano Asymmetric ShuffleBlock for enhanceing feature fusion.
Cross stage partial module with shuffle attention for interference reduction.Processes complex natural backgrounds and lightweight model. [38] 6 V-space-based Multi-scale Feature-fusion SSD Multi-scale attention extremum for automatic lesion detection. Enhances detection ability for disease lesions, especially small ones. [39] 7 LAD-Net Asymmetric and dilated convolution as the convolution to reduce model size.
LAD-Inception designed with an attention mechanism for improving multiscale detection capabilities.Small model size (1.25 MB), high accuracy (97.72%), and implementation of deployment on mobile devices. [40] 8 Enhanced LSTM-CNN Majority voting ensemble classifier replaced the classifier.
Optimal LSTM layer network applied to select deep features autonomously.Enhanced feature extraction and classifier modification. [41] 9 LALNet EARD module with multi-branch structure and depth separable modules extracts more feature information with fewer parameters and computational complexity. SE attention module for increase the feature extraction capability. Small size (6.61 MB), fast execution (6.68 ms/photo), and high recognition accuracy. [42] 10 Two-stage detection system Three-way classification in the first stage using Xception as the base model.
Real-time detection in the second stage.Detects multiple diseases with 87.9% mean average precision. [43] 11 Improved Faster R-CNN Res2Net and feature pyramid network replaced the backbone of Faster R-CNN for batter feature fusion.
RoIAlign instead of RoIPool of Faster R-CNN for improving the identification precision.Extracts multi-dimensional features in natural scenes with complex backgrounds. [44] 12 BC-YOLOv5 Modify YOLOv5 neck structure with weighted BiFPN and CBAM. Enhanced feature extraction in the detection layer, reduced irrelevant information for complex backgrounds. [45] 13 PLPNet Perceptual adaptive convolution (PAC) for enhancing the
network's global sensing capability.
location relation attention module (LRAM) for reducing unnecessary information.
SD-PFAN structure for fusing features batter.Recognizes leaf diseases at the edge of the leaf, resist background interference. [46] 14 DL Technique U-net with Gradient GSO for leaf segmentation in the first stage.
DbneAlexnet trained using proposed GJ-GSO for leaf classification using Gradient Jaya-Golden search optimization in the second stage.Two-stage approach mitigating background noise. Optimized segmentation and classification through new training methods. [47] 15 LightMixer Depth convolution with Phish (DCWP) and light residual (LR) modules to increase feature integration and reduce parameters.
Phish activation function for reducing the information loss.Identifies diseases in complex environments, suitable for mobile deployment. [48] 16 NanoSegmenter Transformer structure and sparse attention mechanisms to tackle the instance segmentation task, replacing the CNN backbone.
The bottleneck inversion technique to achieve model lightweighting.High accuracy in instance segmentation, low computational complexity, and small model size. [49] 17 DMCNN Multi-scale convolution for disease classification from multiple channels. Enhancement of accuracy and efficiency through multi-scale detection [50] 18 CRNN Combines CNN and RNN for improved sequential features extraction. Achieves significant improvement in maximum accuracy compared to traditional CNN. [51] 19 Transfer learning with pre-trained CNN models. Transfer learning with Faster-RCNN and Inception ResNetv2 models. High recognition ability on new dataset after transfer learning. [52] 20 PCA DeepNet Data enhancement with CycleGAN
Feature extraction with PCA
Classification with Faster-RCNN.Innovative PCA method for image extraction, followed by Faster-RCNN for classification. [53] 21 Four transformer-based models. Comparative study on four vision transformers (EANet, MaxVit, CCT, PVT) for tomato leaf disease identification. MaxViT architecture identified as the best for tomato leaf disease identification. [8] 22 Fine-grained image identification framework Utilizes OPM, DRM, AADM, and OCB for object identification, feature learning, and severity assessment. Assess severity based on categorized dataset, captures fine-grained details with DRM. [54] 23 RiceNet YOLOX identifies disease sites in the first stage.
Siamese Network classifies diseases in the second stage.Effective two-stage detection, addressing complex backgrounds and limited samples. [55] 24 RWW-NN SetNet isolates the rice crop images.
RWW algorithm (WWO & ROA), for improved classification.Two-stage approach mitigating background noise, improved classifier performance. [56] 25 The domain adaptation networks with novel attention mechanisms Channel and spatial attention mechanism (CPAM) in DSAN for key feature identification. Alleviates data distribution differences and small sample problems. [57] 26 RiceDRA-Net Res-Attention module based on CBAM for accurate disease identification and localization.
DenseNet-121 serves as the backbone network.Precise disease localization, even in complex backgrounds. [58] 27 rE-GoogLeNet ECA attention mechanism in GoogLeNe
Residual networks for information loss mitigation.Improved recognition and performance over alternatives. [59] 28 ADSNN-BO Enhanced self-attention mechanism employed along the entire architecture in MobileNetV1,
Bayesian optimization for hyperparameter tuning.Outperforms MobileNet with 3.6% accuracy improvement. [60] 29 DGLNet Global attention module (GAM) enhances sensitivity by reducing background noise.
Dynamic representation module (DRM) for flexible feature acquisition.Enhances generalization capability and feature representation in lightweight models. [61] 30 Novel rice grade model EfficientNet-B0 architecture as the backbone for better recognition accuracy for spotting diseases.
By identifying leaf instances and disease areas, the ratio of the two areas was calculated to estimate the severity of the disease.Reliable disease spot recognition, quantifies severity of rice disease. [62] 31 Comparison of pre-trained residual network models Comparison of ResNet34, ResNet50, ResNet18 with self-attention and ResNet34 with self-attention. Models with self-attention exhibit improved recognition accuracy during transfer learning. [63] Table 2.
Leaf disease detection technology and corresponding advantages.
-
Actual Predicted Positive Negative True True positive True negative False False positive False negative Table 3.
Classification of predicted and actual results.
Figures
(1)
Tables
(3)