-
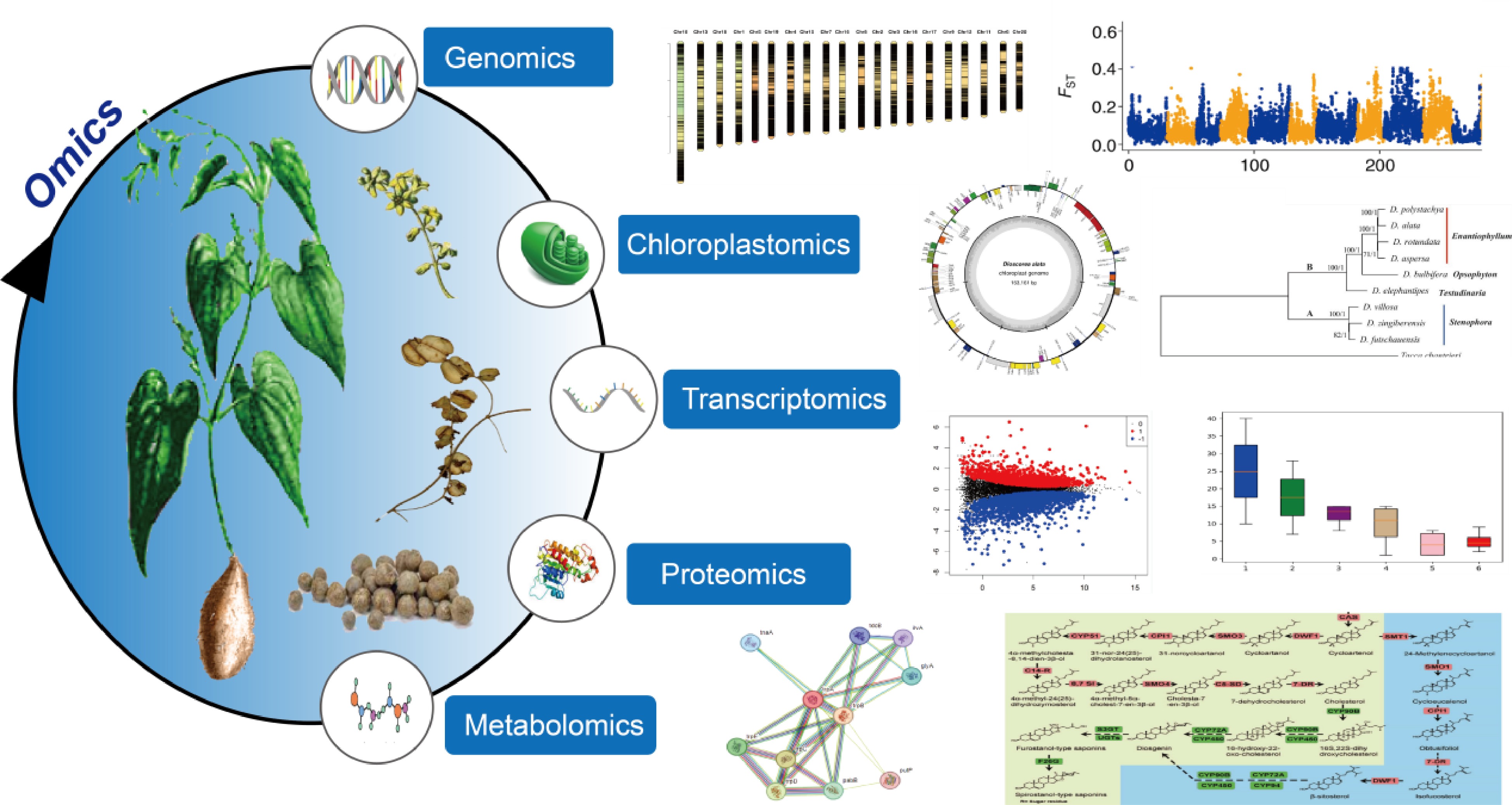
Figure 1.
Omics technologies in yam research.
-
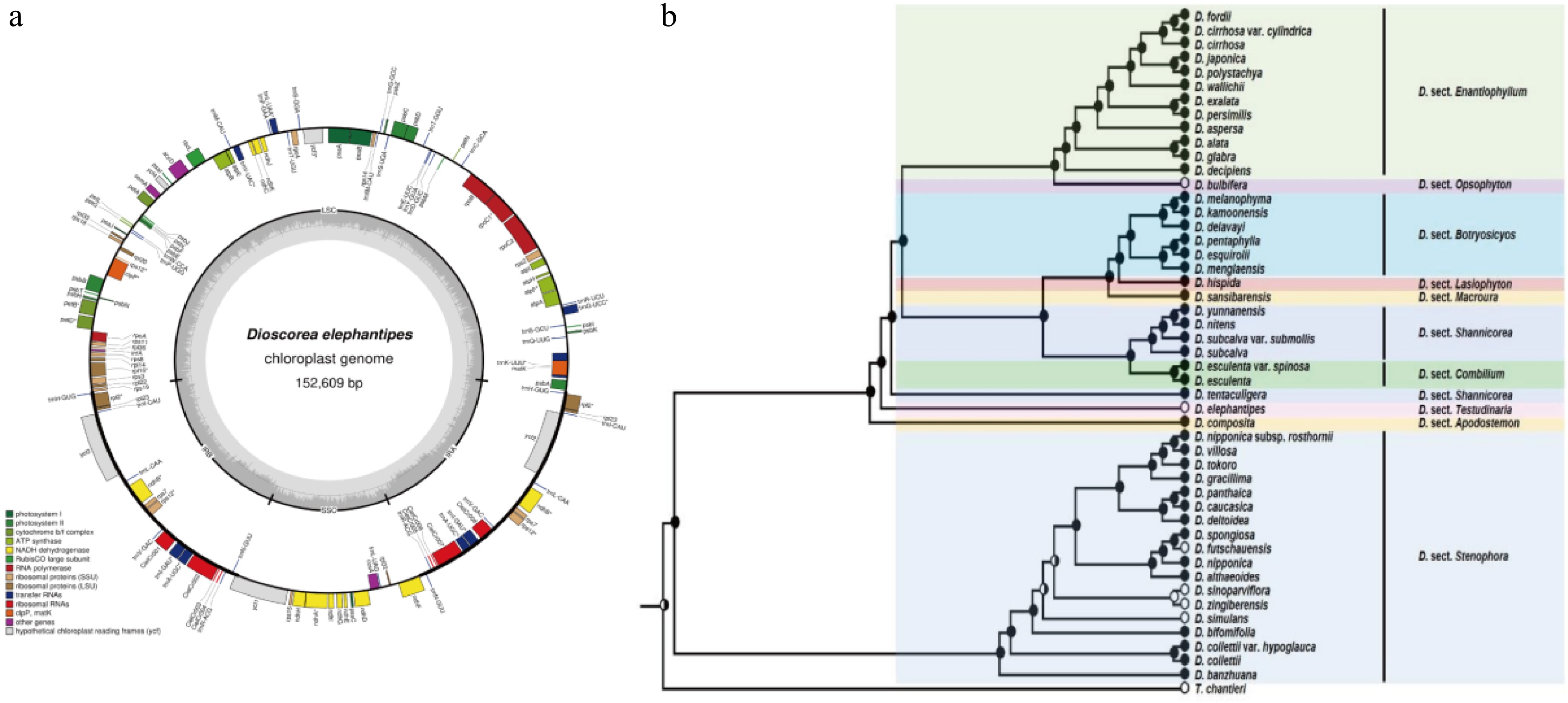
-
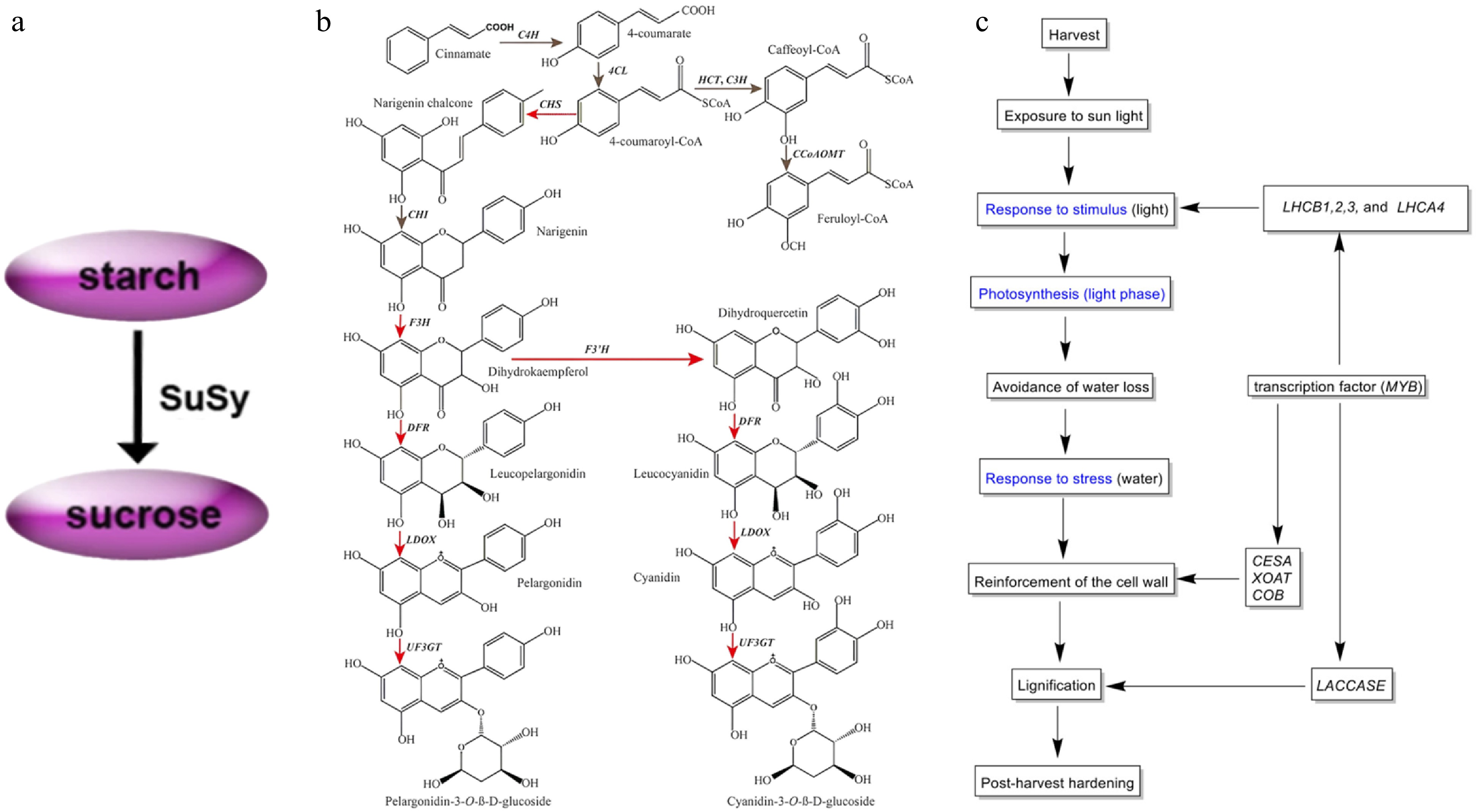
Figure 3.
(a) Modeling the conversion between sucrose and starch during tuber amplification[34]. (b) Simplified diagram depicting the flavonoid biosynthesis pathway in yam tubers. Red arrows indicate genes that were significantly up-regulated in the purple-fleshed yam tuber. Gray indicates no change in gene expression between two tuber types[38]. (c) Putative mechanism of the PHH in D. dumetorum, blue represents GO annotation[41].
-
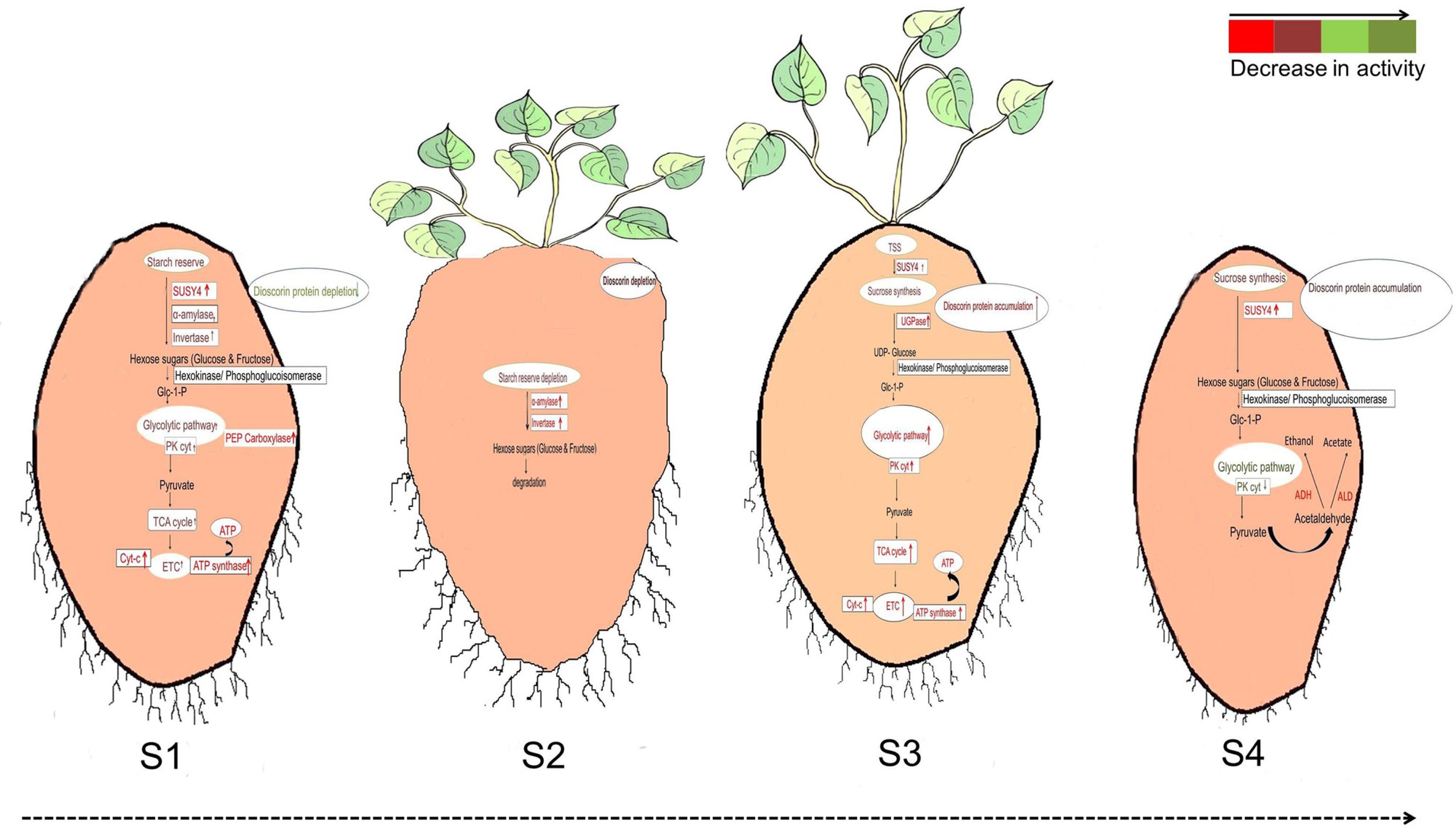
Figure 4.
Differences in the regulation of glycolysis during (S1) tuber sprouting, (S2) degraded tuber, (S3) new tuber formation and (S4) tuber maturation[44].
-
Species Assembly size (Mb) Assembly level N50 (Mb) Gene number Ref. D. rotundata 594 Chromosome 2.12 26,198 [9] D. rotundata* 584 Chromosome 23.4 34,550 [12] D. dumetorum 485 Contig 3.2 35,269 [13] D. alata 479 Chromosome 24 25,189 [3] D. zingiberensis# 480 Chromosome 44.5 26,022 [14] D. zingiberensis# 629 Chromosome 55.8 30,322 [15] D. tokoro 443 Chromosome 24 29,084 [16] * The improved genome assembly for D. rotundata; # The two assemblies from different D. zingiberensis strains. Table 1.
List of sequenced Dioscorea species genomes.
-
Species Metabolites number Germplasm number D. rotundata 99−116 10 D. cayennensis 96−103 4 D. dumetorum 111−130 25 D. alata 104−114 5 D. bulbifera 107−117 5 D. polystachya 431 8 Table 2.
List of mapped Dioscorea species metabolites.
Figures
(4)
Tables
(2)