-
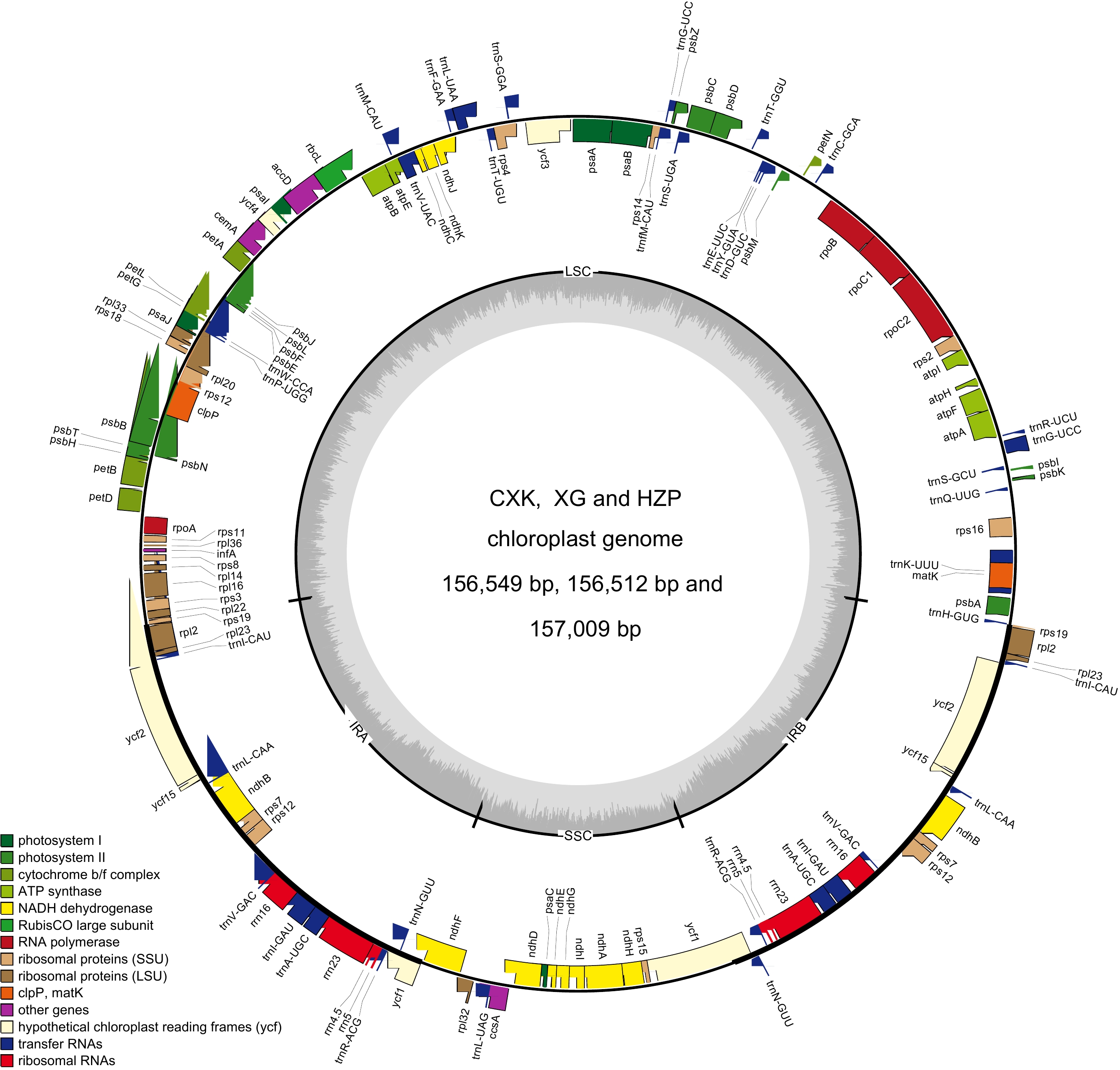
Figure 1.
Chloroplast genome map of C. meiocarpa and C. oleifera.
-
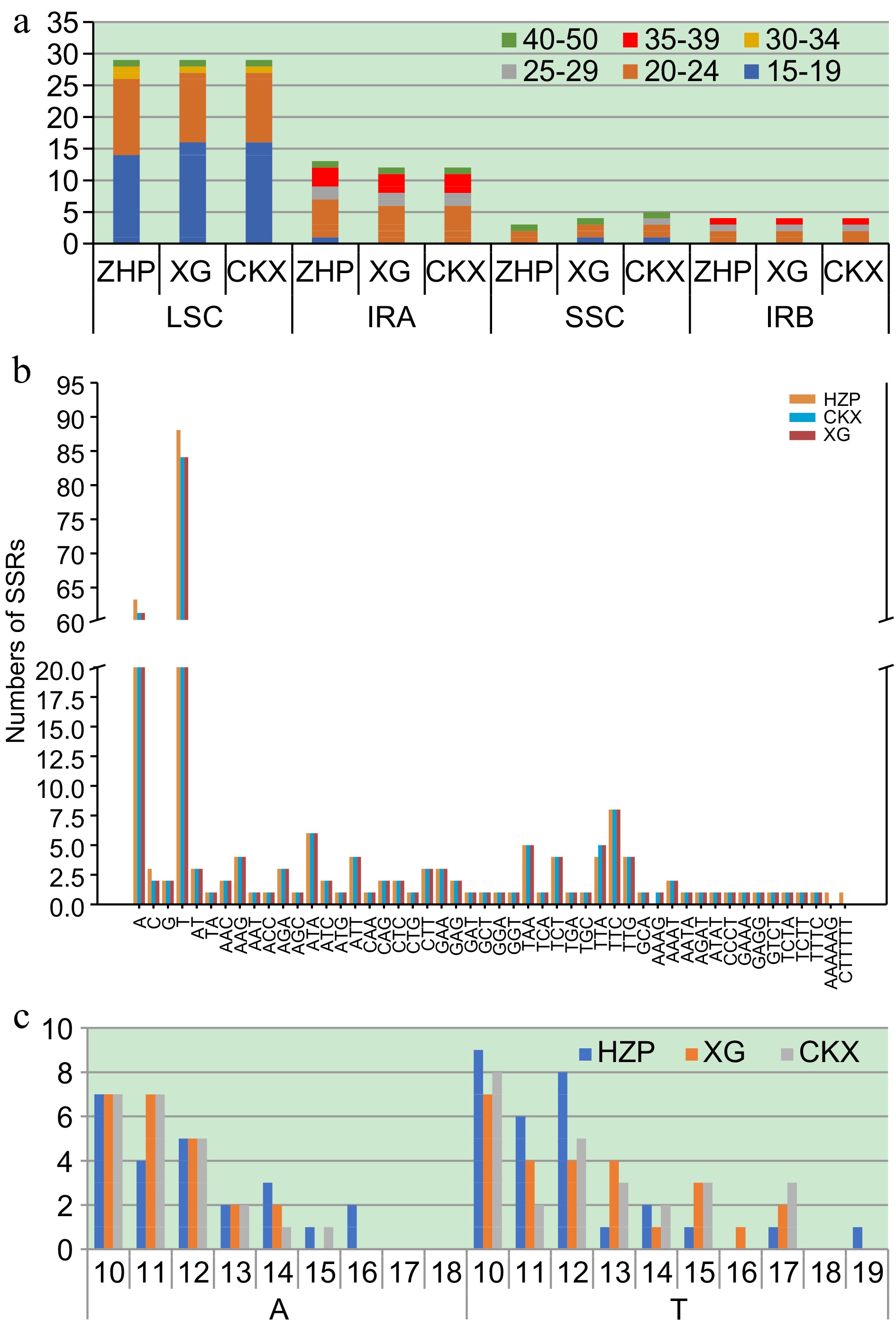
Figure 2.
Comparison of repetitive sequences in C. meiocarpa and C. oleifera. (a) Number of scattered repetitive by length in different regions. (b) The number and distribution of Simple Sequence Repeats (SSR)s. (c) Frequency of SSRs in A and T.
-
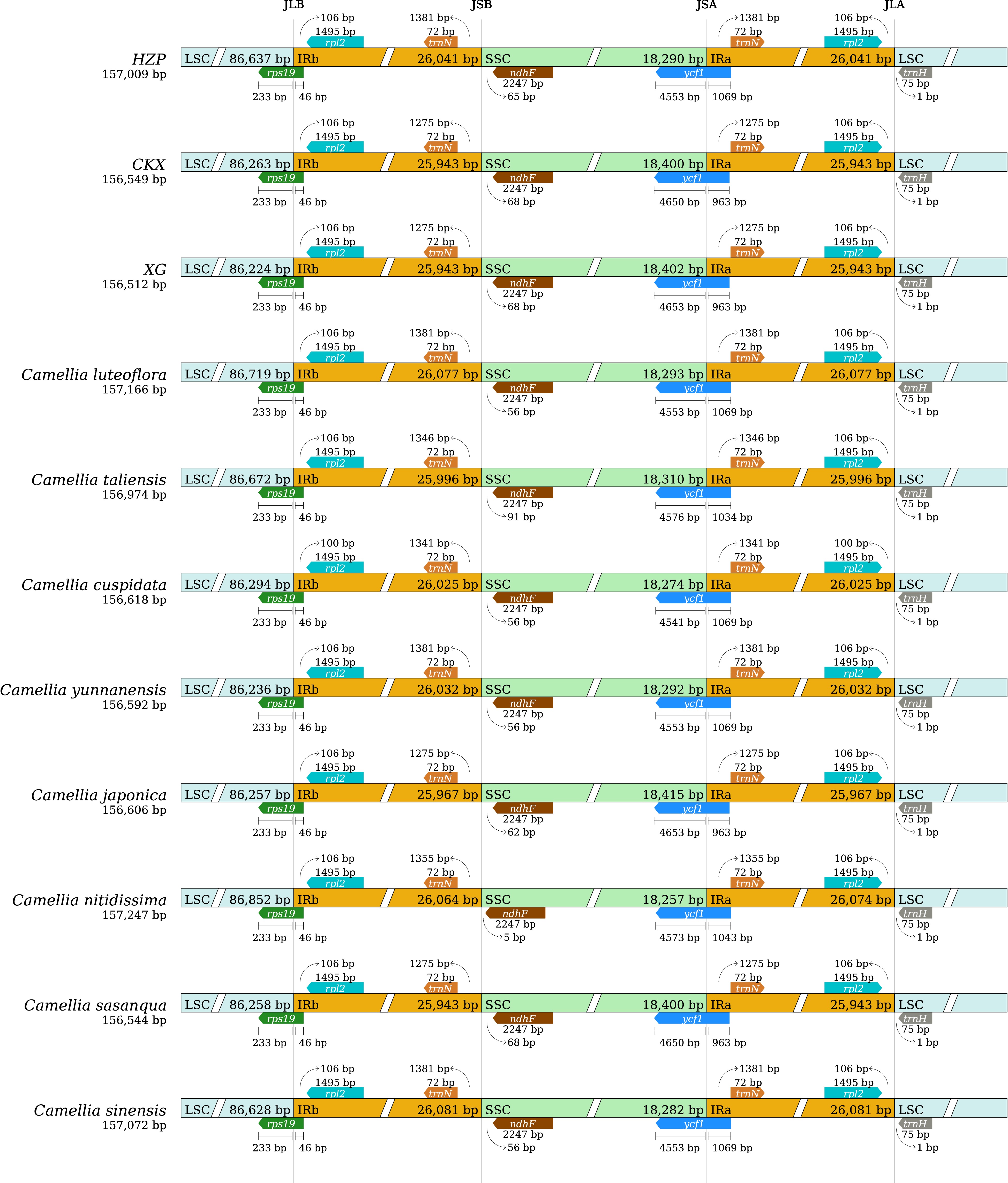
Figure 3.
Comparison of the Large single copy (LSC), Inverted repeat (IR), Small single copy (SSC) junction positions among 11 tea-oil Camellia species.
-
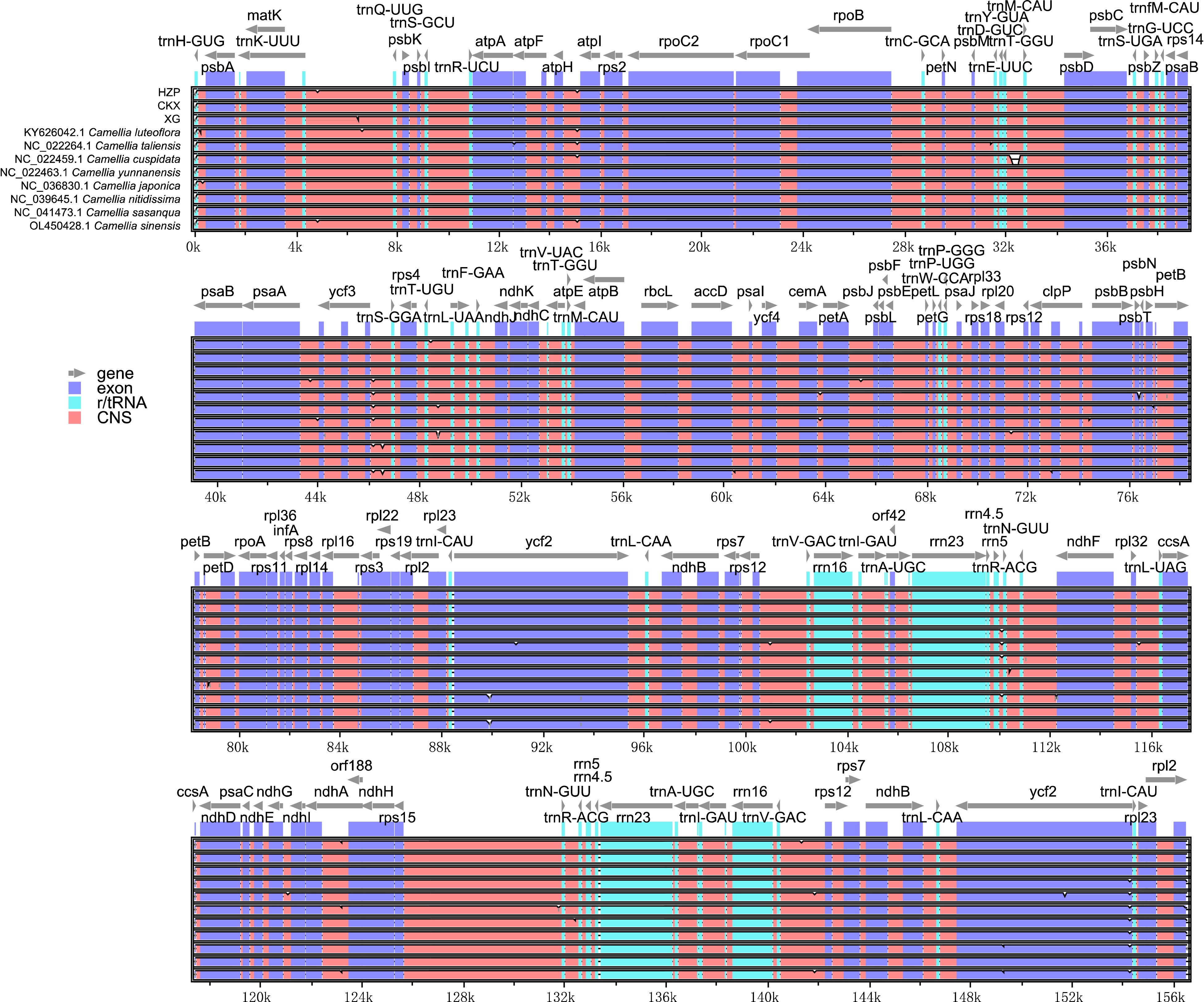
Figure 4.
Identity plots comparing the chloroplast genomes of 11 Camellia accessions. The vertical scale indicates the percentage of identity, ranging from 50 to 100%. The horizontal axis indicates the coordinates within the chloroplast genome. Genome regions are color coded, including protein-coding, rRNA, tRNA, intron, and conserved non-coding sequences (CNS).
-
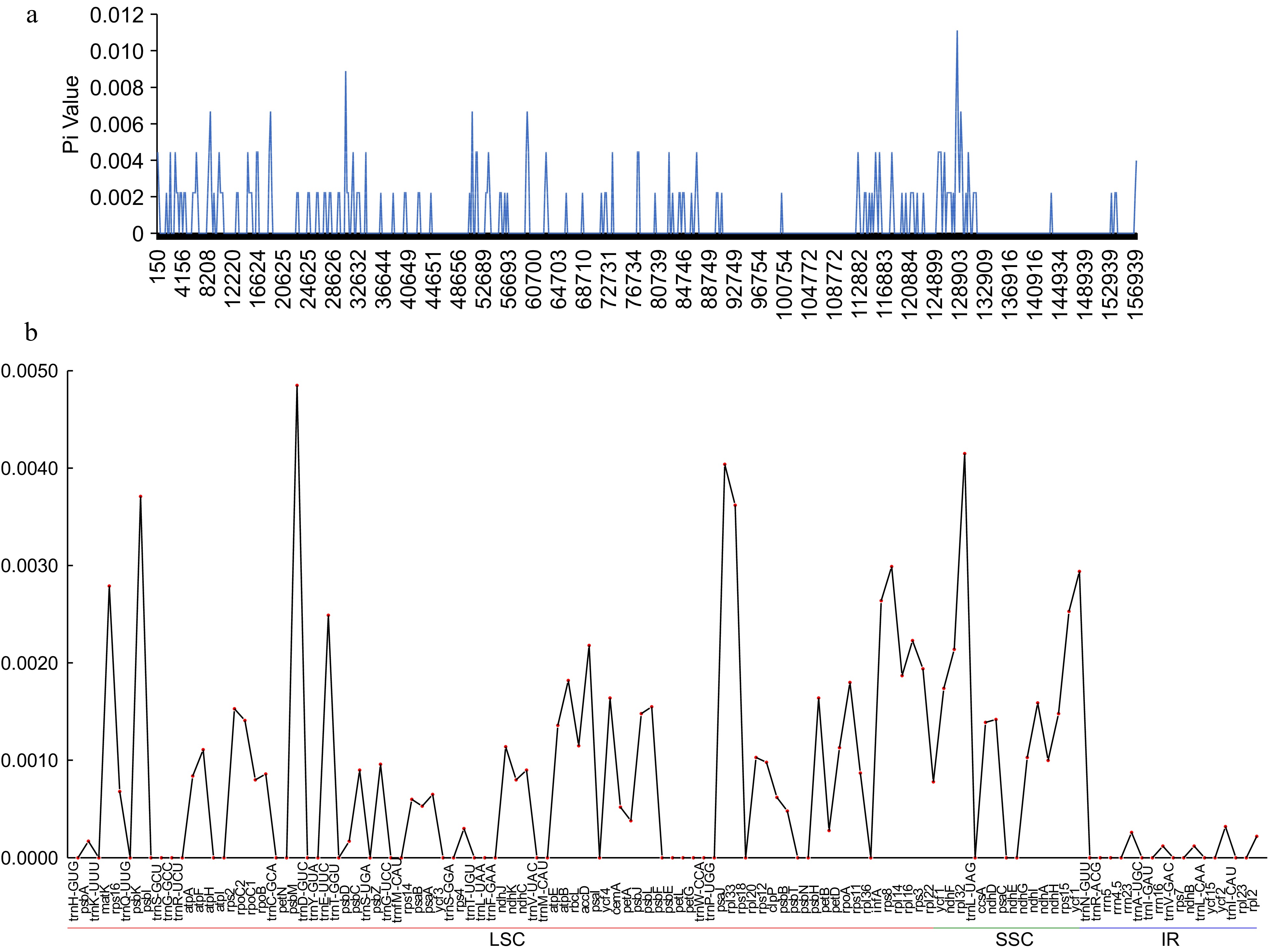
Figure 5.
The nuclear divergence in C. meiocarpa and C. oleifera chloroplast genomes by (a) sliding window analysis of the whole genomes; (b) gene regions.
-
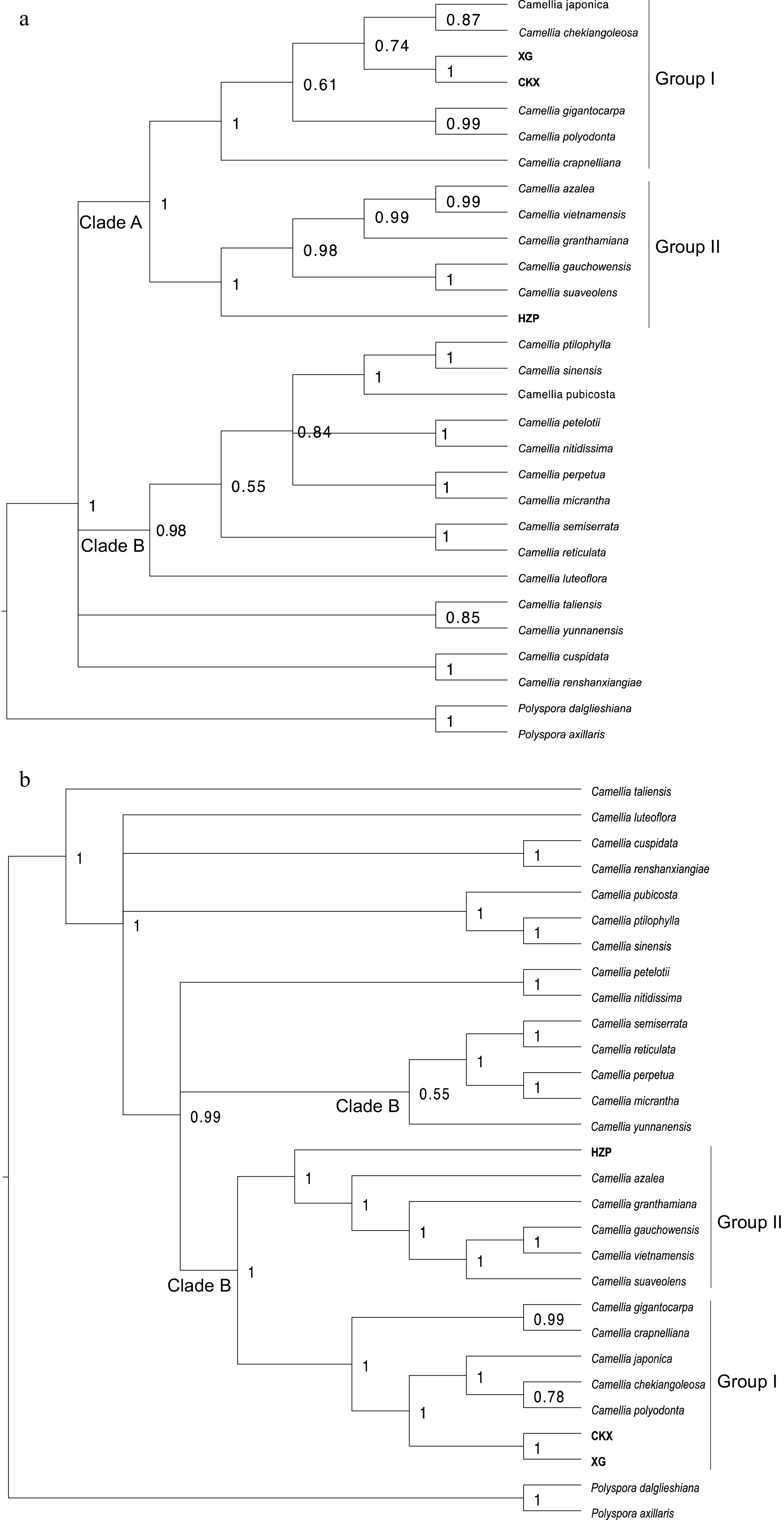
Figure 6.
Phylogenetic tree reconstruction of 27 Camellia species based on (a) protein-coding genes and (b) whole chloroplast genome sequences by the Bayesian method.
-
Genome feature CKX XG HZP Genome size (bp) 156,549 156,512 157,009 LSC length (bp) 86,263 86,224 86,637 SSC length (bp) 18,400 18,402 18,290 IR length (bp) 25,943 25,943 26,041 Number of genes 133 133 133 Number of protien-coding genes 87 87 87 Number of pseudo 2 2 2 Number of tRNA genes 37 37 37 Number of rRNA genes 8 8 8 GC content in LSC (%) 35.33 35.34 35.30 GC content in SSC (%) 30.58 30.57 30.52 GC content in IR (%) 43.03 43.03 42.99 Total GC content (%) 37.32 37.33 37.29 GenBank number MZ151356 MZ151355 MZ151357 Table 1.
Features of C. meiocarpa and C. oleifera chloroplast genomes.
-
ZHP XG CKX Total number 49 49 49 Forward 15 16 17 Palindrome 22 20 20 Reverse 9 9 9 Complementary 3 4 3 Gene N trnS-GCU, trnG-GCC, trnS-UGA, trnfM-CAU,
ndhC, trnV-UACa, petD, ycf2, ndhA, ycf1,
rpoC2b, rpoBc, trnA-UGCdSSR Loci (N) 52 48 49 P1 Loci (N) 51 47 49 Pc Loci (N) 1 1 0 LSC 40 35 36 IRA 2 2 2 SSC 8 9 9 IRB 2 2 2 a: special in ZHP; b, c: special in XG; d: special in CKX. Table 2.
Features of repetitive sequences in C. meiocarpa and C. oleifera.
-
Indel (bp) 1 2 3 4 5 6 9 10−20 21− Total Number (N) 28 12 5 2 10 5 1 6 3 72 Proportion (%) 38.89 16.67 6.94 2.78 1.39 6.94 1.39 8.33 4.17 SNP type G/A C/T A/C G/T C/G T/A Number (N) 36 32 27 27 5 11 138 Proportion (%) 26.09 23.19 19.57 19.57 3.62 7.97 Table 3.
Indel and SNP types among three chloroplast genomes.
-
Primers Loci SNP Indel ZDJ01 TCCACTATTT[C/A]AATTATAAAA 1 0 ZDJ01 CAACCCATAA[C/-]CCATAAAAAT 0 1 ZDJ03 CCCAAAAAAT[G/A]GATTTTGGTT 1 0 ZDJ15 TCAATGGCCC[T/C]CCTACGTAGT 1 0 ZDJ45 TCCCATATAT[T/-]AAATATTAAA 0 1 ZDJ51 ATTGAAAGCT[A/G]GGATTTCTAG 1 0 ZDJ54 AATCCTTGTT[T/G]CGGAGTCGAT 1 0 ZDJ55 ACCAAAAAAT[A/C]TTTTTTGCTT 1 0 ZDJ59 TTCATCTATT[T/C]CATGACCGGA 1 0 ZDJ60 GACCAAGAAG[G/-]ATTCTCTTTC 0 1 ZDJ69 ATAAAAAATT[A/T]CCCCCTGCAA 1 0 ZDJ72 AAAATCATGT[G/A]TTGGTCCAGA 1 0 ZDJ76 TTCAAAATGG[C/-]TTTCAAATTA 0 1 ZDJ76 AAAGAATAGT[A/C]AATTTTTGCA 1 0 ZDJ76 AGAATAATTT[G/T]AATCTTAAAA 1 0 ZDJ77 GTATAACCCC[C/T]TTTTGCTTTC 1 0 ZDJ80 TAAGAATGGG[G/T]GACGGTATTC 1 0 ZDJ83 GAATTCTGTG[A/G]AAAGCCGTAT 1 0 ZDJ84 AAGAGAATCC[T/-]TCTTGGTCGT 0 1 ZDJ85 TCCGGTCATG[A/G]AATAGATGAA 1 0 In Loci, the variant in left side was C. oleifera and the right side was C. meiocarpa. Table 4.
The SNP and Indel in the targeted regions.
Figures
(6)
Tables
(4)