-
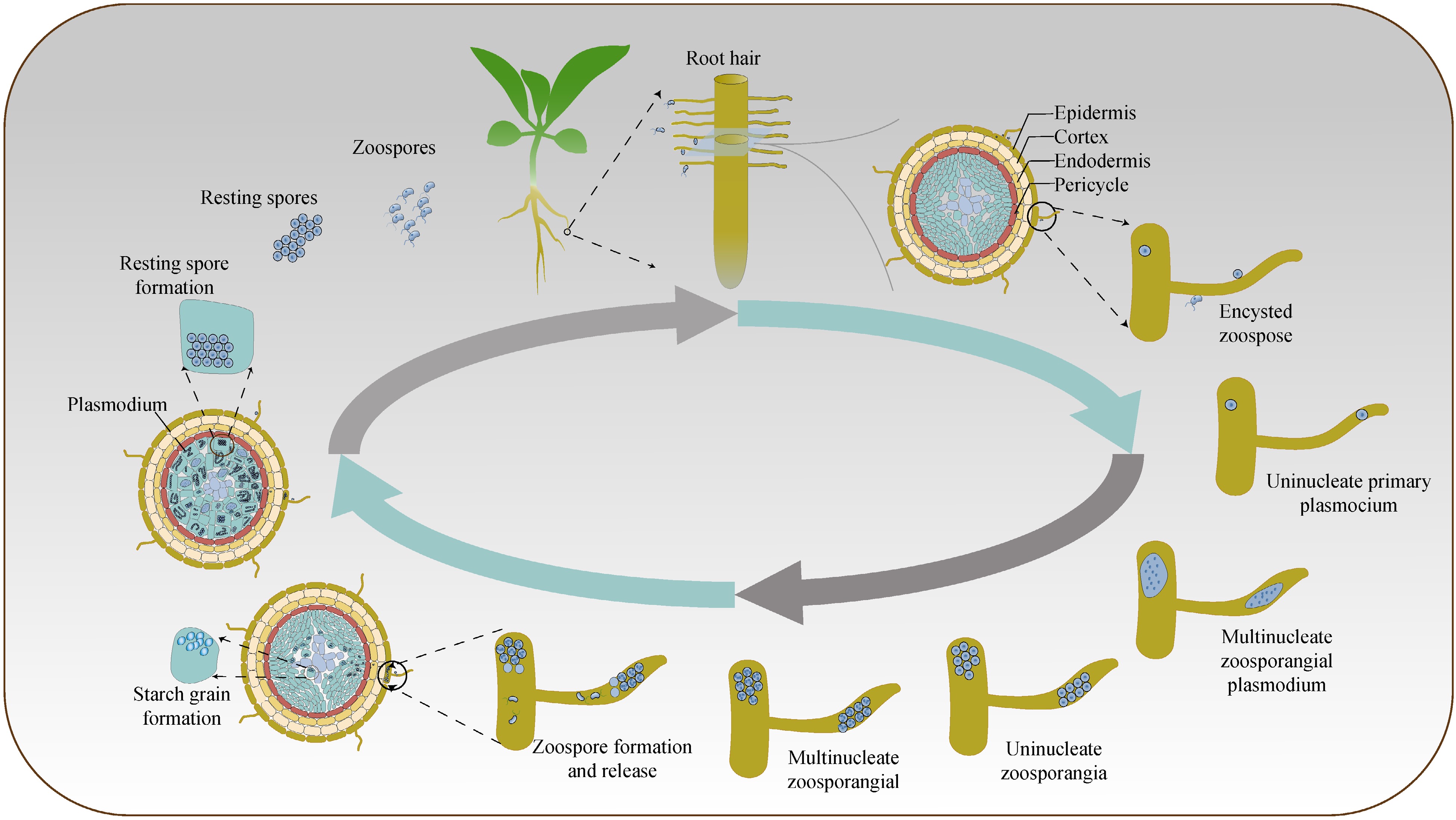
Figure 1.
Diagram of the refined life cycle of Plasmodiophora brassicae.
-
Pathotype classification systems Host (cultivar number) Original strain No. Races/pathotypes Ref. Williams' classification system B. oleracea (2) and B. napobrassica (2) − 16 [24] European clubroot differential (ECD) B. rapa (5), B. napus (5), and B. oleracea (5) − 34 [27] Somé system B. napus (10) 20 5 [30] Kuginuki system B. rapa (10), B. napus (2), B. oleracea (2), and B. napobrassica (2) 36 − [31] Canadian clubroot differential (CCD) B. rapa (2), B. napus (9), and B. oleracea (2) 106 17 [32] Sinitic clubroot differential (SCD) B. rapa (7), B. oleracea (2), and B. napobrassica (2) 132 16 [33] Table 1.
Summary for classification systems of P. brassicae pathogens.
-
Species CR gene/QTL Linked markers Plant material Mapping population Pathogen race Gene position Ref. B. rapa L. CRa HC352b-SCAR R: DH line 'T136-8' F2 M85 (Race2) A03 [92,100] S: DH line 'Q5' &'K10' CRaim-T R: DH line 'T136-8' S: DH line 'Q5' Crr1 BRMS-088 R: DH line 'G004'
S: DH line 'A9709'F2 Wakayama-01 A08 [101] Crr2 BRMS-096, BRMS-100 R: DH line 'G004'
S: DH line 'A9709'F2 Wakayama-01 A01 [101] Crr3 BrSTS-41, BrSTS-54 R: Inbred line 'NWMR-3 ' F2 Ano-01 A03 [101,102] S: DH line 'A9709 ' Crr4 R: DH line 'G004' F2 Wakayama-01 and Ano-01 A06 [103] S: DH line 'A9709' CRb TRC05, TRC09 R: DH line 'CRShinki' F2 Race 4 A03 [104] S: DH line '94SK' K-3 R: Inbred line 'CCR13685' F2, BC1 Race 4 [105] S: Inbred line 'GHQ11021' TCR79, TCR108 R: DH line 'CRShinki' F2 Pathotype 4 [10] S: Inbred line '702-5' SC2930-Q-FW/SC2930-RV R: DH lines 'T136-8','K13'
S: Inbred line '702-5'F2 M85 (Race2) K04 [106] SC2930-T-FW/SC2930-RV CRc B50-C9-FW/B50-RV R: DH lines 'C9', 'RC22' F2 M85 (Race2) K04 A02 [92,107] B50-6R-FW/B50-RV S: DH line 'Q5' CRk HC688-4-FW/HC688-6-RV A03 [92] HC688-4-FW/HC688-7-RV Crr1a
Crr1bBSA7
BSA2R: DH lines 'G004'
S: DH line 'A9709'F2, BC3F3 Ano-01, Wakayama-01 A08
A08[94] PbBa3.1 sau_um438a R: Turnip 'ECD04'
S: Chinese cabbage 'C59-1'BC1F1 Pb2 A03 [108] PbBa3.3 sau_um398a Pb7 A03 [108] PbBa8.1 cnu_m490a Pb4 A08 [108] Rcr1 (Rpb1) MS7-9 R: Hybrid cv. FN
S: Canola DH line 'ACDC'F1 Leduc-AB-2010 A03 [109] QS_B1.1 BRMS287-aaf SN3523a R: Inbred line 'Siloga'
S: Inbred line 'BJN3'F2 Wakayama-01 A01 [110] QS_B3.3 sau_um028-At4g35530 A03 QS_B8.1 BRPGM0920-BRPGM0173 A08 Rcr4 A03_23710236 R: Canola 'T19'
S: DH line 'ACDC'F2:3, BC1S1 Pathotype 2 A03 [111] Rcr8 A02_18552018 Pathotype 5× A02 Rcr9 A08_10272562 A08 Rcr2 SNP_A03_08 R: Chinese cabbage 'Jazz' BC1S1 Pathotype 3 A03 [112] SNP_A03_09 S: DH line 'DHACDC' A03 CRd yau389, yau376 R: Inbred line '85-74'
S: Inbred line 'BJN3-1'F2:3 Pathotype 2, 4, 7, and 11 A03 [113] CrrA5 TuuYCBRCR404 R: Inbred line '20-2cc1'
S: Inbred line 'ЕС-1'BC1 A05 [114] CRs R: Turnip 'SCNU-T2016'
S: Cabbage 'CC-F920'F2:3 Group 4 A08 [115] Rcr3 A90_A08_SNP_M11 R: '96-6990-2' F1, BC1 Pathotypes 3H and 5X A08 [91] Rcr9wa A90_A08_SNP_M28 S: DH line 'ACDC' A08 BraA3P5G.CRa/bKato1.1 KB59N06, B4732 R: Inbred line 'ECD 02'
S: 'CR 2599', 'CR 1505'F2 Pathotype 2B, 2F, 3A, 3D, 3H, 3O, 5C, 5G, 5I, 5K , 5L, 5X, 6M, 8E, 8J, 8N, and 8P A03 [116] BraA3P5G.CRa/bKato1.2 CRaJY, BGB41 A03 [116] Bcr1 A03-1-192 R: Inbred line '877' F2 A03 [89] Bcr2 A03-1-024 S: Inbred line '255' A08 [89] CRq Br-insert1 R: DH lines 'Y635-10'
S: DH line 'Y177-47'F2 A03 [117] CRA8.1 A08-4346 R: 'H5R' and '409R' F1 PbXm, PbCd, PbZj, PbTc, and PbLx A08 [118] A08-4624 S: 'H5S' and '91-12' CRA3.7 syau-InDel3008 R: Inbred line 'CR510' F2 Pb3 A03 [119] S: Inbred line '59-1' B. oleracea L. CR2a R: 'No. 86-16-5' F2:3 Race 2 LG6 [120] CR2b S: 'CrGC No. 85' LG1 [120] pb-3 4NE11a R: DH line 'Bi' F2:3 ECD16/3/30 LG3 [121] pb-4 2NA8c S: DH line 'Gr' LG1 Pb-Bo1 T2 R: Inbred line 'C10' F2:3 P1 (Ms6 and eH), P2 (K92), P4 (K92-16) and P7 (Pb137-522) LG1 [122] Pb-Bo2 s07.1900 S: DH line 'HDEM' LG2 Pb-Bo3 aa7.1400 LG3 Pb-Bo4 aa9.983 LG4 Pb-Bo5a PBB7b LG5 Pb-Bo5b a18.1400 LG5 Pb-Bo8 c01.980 LG8 Pb-Bo9a aj16.570 LG9 Pb-Bo9b a04.1900 LG9 QTL1 CA69b, CB85a R: Inbred line 'K269'
S: Cabbage line 'Y2A'F2:3 Kamogawa, Anno and Yuki O3 [123] QTL3 CA63 O3 QTL9 CA93 O3 pb-Bo(Anju)1 KBrH059L13 R: DH line 'Anju'
S: DH line 'GC'F2:3 Race 4 O2 [124] pb-Bo(Anju)2 m6R O2 pb-Bo(Anju)3 BRMS-330 O3 pb-Bo(Anju)4 KBrS012D09N1 O7 pb-Bo(GC)1 ACTb, CB10435 O5 CRQTL-GN_1 C2h-1(4), C2h-5(4) R: Inbred line 'C1220'
S: Inbred line 'C1176'F2, F2:3 Race 2, Race 9 O2 [125] CRQTL-GN_2 C3b-3(8), C3a-34(2) O3 CRQTL-YC C3a-65(8) O3 Rcr_C01-1 D134_C01_8,398,944 R: Inbred line 'ECD11'
S: DH line 'T010000DH3'BC1/BC1S1 Pathotype 3A, 2B, 5C, 3D, 5G, 3H, 8J, 5K, 5L and 3O C01 [126] Rcr_C03-1 D134_C03_9,211,088 C03 Rcr_C03-2 D134_C03_585,685 C03 Rcr_C03-3 D134_C03_35,229,606 C03 Rcr_C04-1 D134_C04 _51,280,226 C04 Rcr_C08-1 D134_C08_23,354,593 C08 Rcr_C08-2 D134_C08_28,507,471 C08 B. napus L. Pb-Bn1 OPG03.960 R: DH line 'Darmor-bzh' F1 Pathotypes
4 and 7LG 4 [127] S: Inbred line 'Yudal' PbBn_di_A02 BS008863 Partially resistant 'Aviso' and 'Montego' DH Pathotype P1 A02 [128] PbBn_di_C03 BS006202 C03 PbBn_di_C04 BS007532 C04 PbBn_rsp_C03 BS012716 C03 qCR_A8 Bn-N3-p16098951 R: 'Rutabaga-BF' DH Pathotypes 2, 3, 5, 6 and 8 A08 [129] qCR_A3 UACSSR3667 S: 'UA AlfaGold' A03 Rcr10ECD01 DM_A03_12570715 R: Inbred line 'ECD01' F1, BC1 Pathotypes 3A, 3D, and 3H A03 [130] Rcr9ECD01 DM_A08_10325589 S: Inbred line 'DH16516' A08 ERF034 BnSNP14198336 R: Inbred line 'Kc84R' F2 Pathotypes 2, 4, 7, and 11 A03 [131] S: Inbred line '855S' R. sativus L. Crs1 RSACCCTC4 R: 'Utsugi-gensuke' F2 Ano-01 and Wakayama-01 LG1 [132] REL24, REL6 S: 'Koga benimaru' RsCr1 R09_11227501 R: Inbred line 'BJJ'
S: Inbred line 'XNQ'F2 Pb10 R09 [133] RsCr2 R09_11933628 R09 RsCr3 R09_15947806 R09 RsCr4 R08_16258481 R08 RsCr5 R08_26984449 R08 RsCr6 HB321, HB331 R: Inbred line 'GLX' F2 Pb10 R08 [134] S: Inbred line 'XNQ' B.nigra L. Rcr6 SNP_B03_51, R: 'PI 219576' F1, BC1, F2 Pathotype 3 B07 [135] SNP_B03_52 S: 'CR2748' Table 2.
CR genes, linked markers, and QTLs in plant species of the Brassicaceae family.
Figures
(1)
Tables
(2)