-
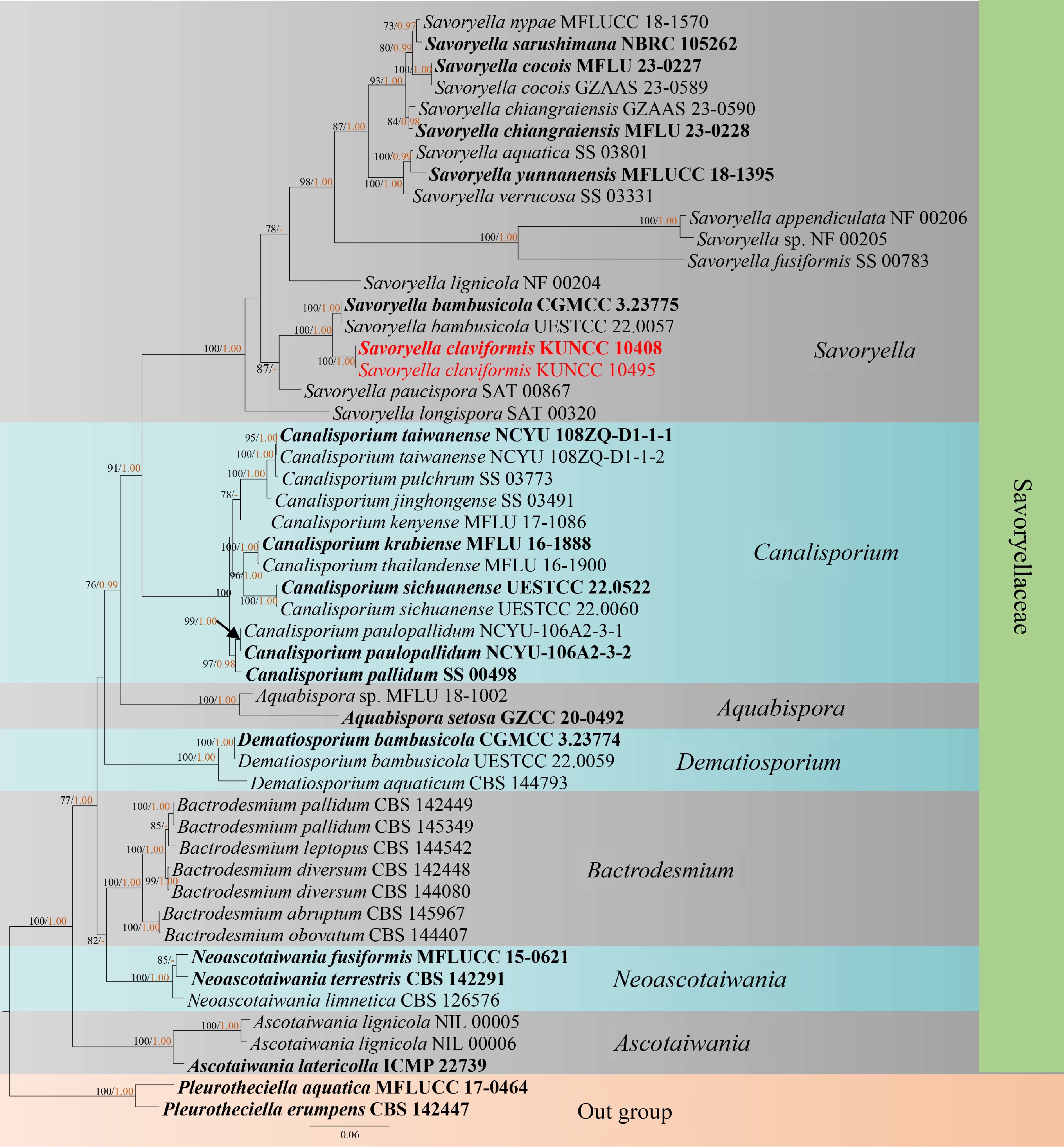
Figure 1.
RAxML tree based on analysis of a combined LSU, SSU, and ITS sequences for Savoryellaceae. Bootstrap support values for maximum likelihood (ML) equal to or greater than 75% were given above the nodes (left). Bayesian posterior probability (BIPP) equal to or greater than 0.95 were given above the nodes (right) and hyphen (−) were marked as values below 0.95. The tree was rooted to Pleurotheciella aquatica (MFLUCC 17-0464) and P. erumpens (CBS 142447)[18]. Two new isolates were shown in red, and ex-type strains are bold.
-

Figure 2.
Savoryella claviformis (HKAS 133191, holotype). (a) Colony on nature substrates. (b), (c) Conidiophores and apical conidia. (d) Conidiophore. (e)–(g) Conidia. (h) Culture colony on PDA medium, from surface. (i) Culture colony on PDA medium, from reverse. Scale bars: (b)–(g) = 50 μm.
-
Taxa Vouchers/strains/isolates GenBank accession numbers ITS LSU SSU Aquabispora sp. MFLU 18-1002 MK421951 MK421953 MK421952 Aquabispora setosa GZCC 20-0492 OP377819 OP377918 OP378003 Ascotaiwania latericolla ICMP 22739 MN699390 MN699407 – Ascotaiwania lignicola NIL 00005 HQ446341 HQ446364 HQ446284 Ascotaiwania lignicola NIL 00006 HQ446342 HQ446365 HQ446285 Bactrodesmium abruptum CBS 145967 MN699393 MN699410 MN699367 Bactrodesmium diversum CBS 142448 MN699352 MN699412 MN699369 Bactrodesmium diversum CBS 144080 MN699355 MN699415 MN699371 Bactrodesmium leptopus CBS 144542 MN699388 MN699423 MN699374 Bactrodesmium obovatum CBS 144407 MN699397 MN699426 MN699377 Bactrodesmium pallidum CBS 142449 MN699363 MN699428 MN699379 Bactrodesmium pallidum CBS 145349 MN699364 MN699429 MN699380 Canalisporium jinghongense SS 03491 GQ390287 GQ390272 GQ390257 Canalisporium kenyense MFLU17-1086 MH701998 MH701999 – Canalisporium krabiense MFLU 16-1888 MH275051 MH260283 – Canalisporium pallidum SS 00498 GQ390295 GQ390280 GQ390265 Canalisporium paulopallidum NCYU-106A2-3-1 MT946658 – – Canalisporium paulopallidum NCYU-106A2-3-2 MT946659 – – Canalisporium pulchrum SS 03773 GQ390293 GQ390278 GQ390263 Canalisporium sichuanense CGMCC 3.23926 OQ428270 OQ428262 OQ428254 Canalisporium sichuanense UESTCC 22.0060 OQ428271 OQ428263 OQ428255 Canalisporium taiwanense NCYU-108ZQ-D1-1-1 MT946663 – – Canalisporium taiwanense NCYU-108ZQ-D1-1-2 MT946664 – – Canalisporium thailandense MFLU 16-1900 MH275052 MH260284 – Dematiosporium aquaticum CBS 144793 MN699402 MN699433 MN699385 Dematiosporium bambusicola CGMCC 3.23774 OQ428268 OQ428260 OQ428252 Dematiosporium bambusicola UESTCC 22.0059 OQ428273 OQ428265 OQ428256 Neoascotaiwania fusiformis MFLUCC 15-0621 MG388215 KX550893 – Neoascotaiwania limnetica CBS 126576 KY853452 KY853513 KT278689 Neoascotaiwania terrestris CBS 142291 KY853454 KY853515 KY853547 Pleurotheciella aquatica MFLUCC 17-0464 MF399236 MF399253 MF399220 Pleurotheciella erumpens CBS 142447 MN699406 MN699435 MN699387 Savoryella appendiculata NF 00206 HQ446350 – HQ446293 Savoryella aquatica SS 03801 HQ446349 HQ446372 HQ446292 Savoryella bambusicola CGMCC 3.23775 OQ428269 OQ428261 OQ428253 Savoryella bambusicola UESTCC 22.0057 OQ428267 OQ428259 OQ428251 Savoryella claviformis KUNCC 10408 OP626331 PP577958 PP577960 Savoryella claviformis KUNCC 10495 PP580830 PP577959 PP577961 Savoryella fusiformis SS 00783 HQ446351 – HQ446294 Savoryella lignicola NF 00204 HQ446357 HQ446378 HQ446300 Savoryella longispora SAT 00320 HQ446358 HQ446379 HQ446301 Savoryella nypae MFLUCC 18-1570 MK543219 MK543210 MK543237 Savoryella paucispora SAT 00867 HQ446361 HQ446382 HQ446304 Savoryella sarushimana NBRC 105262 – MK411004 MK411005 Savoryella verrucosa SS 03331 HQ446355 HQ446376 HQ446298 Savoryella yunnanensis MFLUCC 18-1395 – MK411422 MK411423 Savoryella sp. NF 00205 HQ446362 – HQ446305 Savoryella cocois MFLU 23-0227 OR581911 OR438867 OR458366 Savoryella cocois GZAAS 23-0589 OR581912 OR438868 OR458367 Savoryella chiangraiensis GZAAS 23-0590 OR581914 OR438870 OR458369 Savoryella chiangraiensis MFLU 23-0228 OR581913 OR438869 OR458368 The newly generated sequences are indicated in blue. The ex-type strains are in bold and '–' indicates unavailable sequences. Table 1.
Taxa used in the phylogenetic analyses and their corresponding GenBank accession numbers.
Figures
(2)
Tables
(1)