-
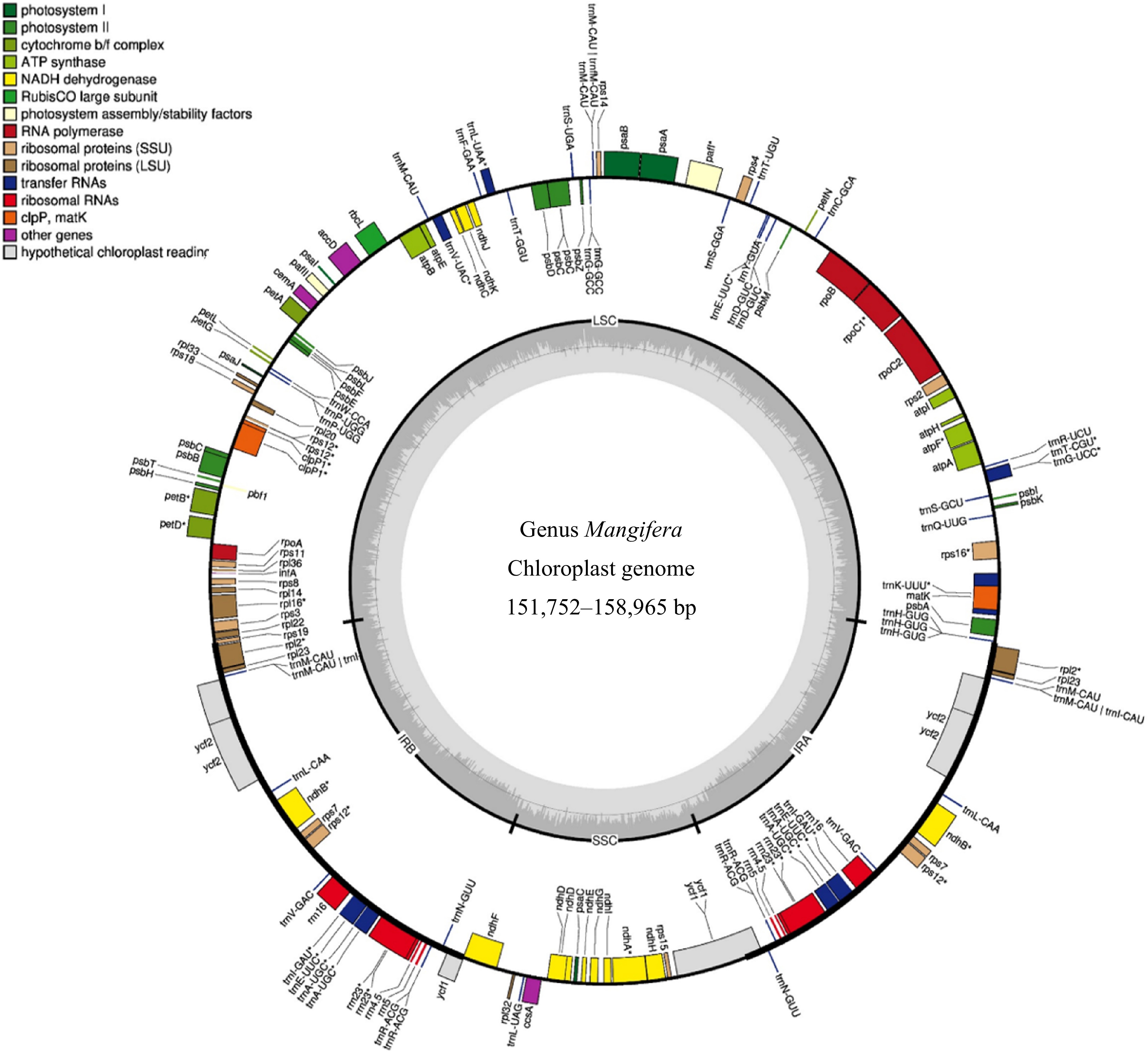
Figure 1.
Genome map of the chloroplasts in the genus Mangifera. The genome size of the 16 Mangifera species ranges from 151,752 to 158,965 bp for M. caesia and M. laurina, respectively. In the most outer circle, the thick black border/line indicates Inverted Repeat Regions (IR) whereas the thin lines indicate Large Single Copy (LSC) and the Small Single Copy (SSC). Genes inside the circle are transcribed in the clockwise direction whereas the genes outside the circle are transcribed in the counter-clockwise direction. Different colours are given for the genes with respect to their functions. The darker grey in the inner circle corresponds to GC content, whereas the lighter grey corresponds to AT content.
-
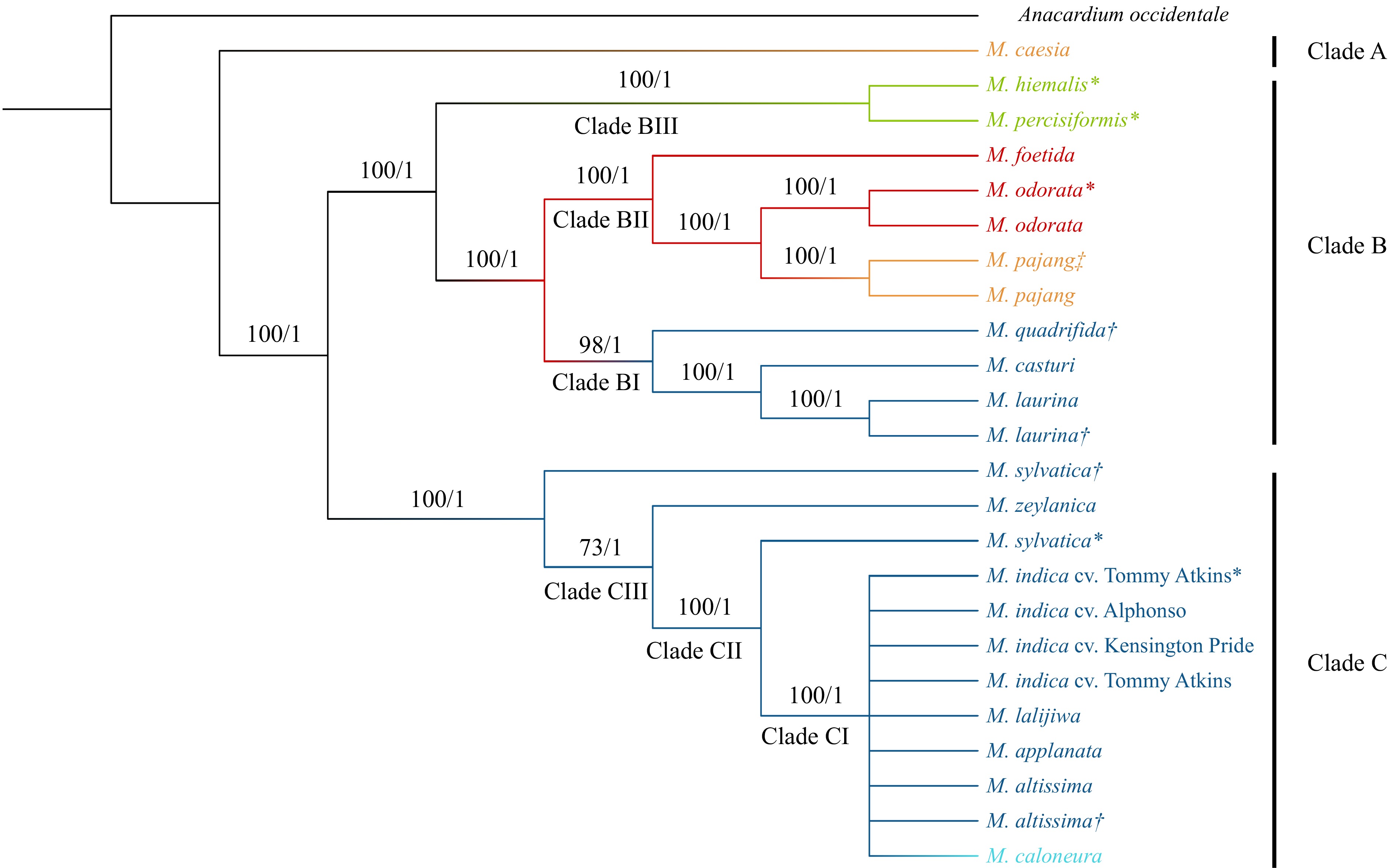
Figure 2.
Phylogenetic tree developed for Mangifera species based on whole chloroplast genomes. The phylogenetic tree of 24 accessions belongs to 16 species with A. occidentale used as the outgroup. Trees were generated using Maximum Likelihood (ML) and Bayesian inference (BI) method. Numbers associated with the branches are ML bootstrap value (/100) and BI posterior probabilities (/1). Dark Blue: Sub genus Mangifera, Section Mangifera, Light blue: Sub genus Mangifera, Section Euantherae, Red: Sub genus: Limus, Section Perrennis, Yellow: Sub genus: Limus, Section: Deciduae, Light Green: species placed in uncertain position in the classification. * Mangifera species for which chloroplast genomes were assembled using raw data downloaded from NCBI. † Mangifera species collected from the Botanical Ark, Queensland, Australia. ‡ Mangifera species collected from the Durian Heaven Farm, Queensland, Australia.
-
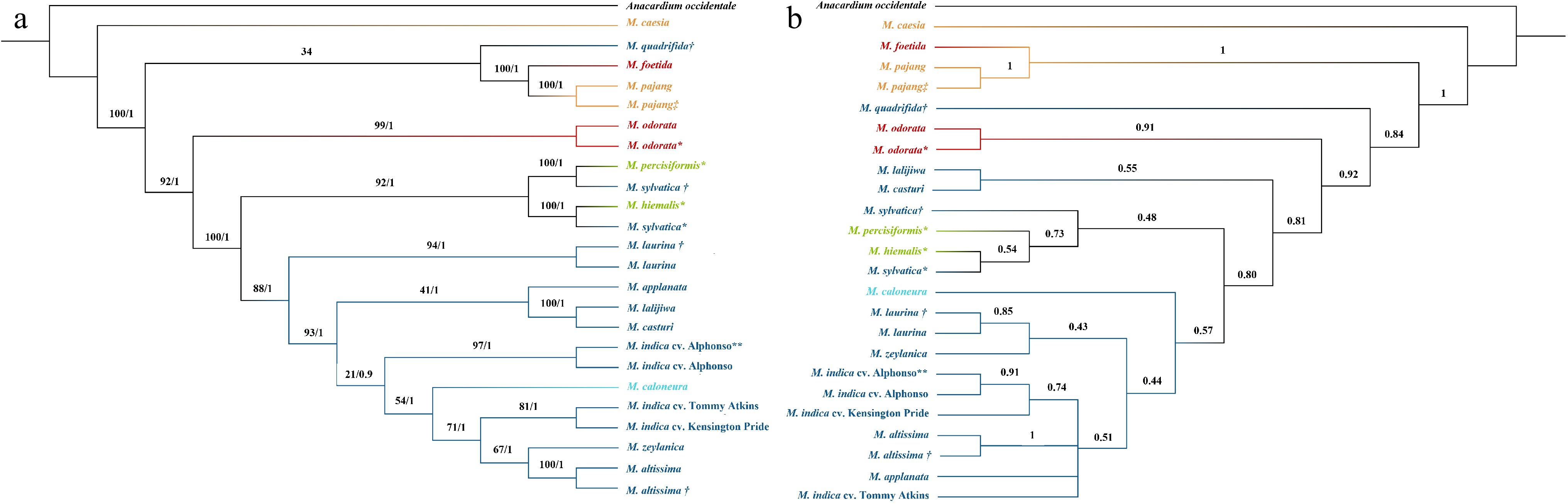
Figure 3.
Phylogenetic tree developed for Mangifera species based on a selected set of nuclear genes using (a) concatenation and (b) coalescence-based methods. Phylogenetic tree of 24 accessions with A. occidentale used as the outgroup. Concatenation-based trees were generated using Maximum Likelihood (ML) and Bayesian inference (BI) methods and consensus tree is shown in the figure. Numbers associated with the branches are ML bootstrap value (/100) and BI posterior probabilities (/1). In the coalescence-based tree (ASTRAL tree), numbers associated with branches are local posterior probability values (/1). Dark Blue: Sub genus Mangifera, Section Mangifera, Light blue: Sub genus Mangifera, Section Euantherae, Red: Sub genus: Limus, Section Perrennis, Yellow: sub genus: Limus, Section: Deciduae. * Species for which nuclear genes were extracted using raw data downloaded from NCBI. ** M. indica cultivar from which gene models were downloaded from NCBI and used to create local database in CLC-GWB for the selection of single copy nuclear genes in M. indica. † Mangifera species collected from the Botanical Ark, Queensland, Australia. ‡ Mangifera species collected from the Durian Heaven Farm, Queensland, Australia.
-
Sub genera Section Species/taxon Embryony Country of origin Geographical distribution of the species Ploidy level (2n) Prominent fruit/horticultural characteristics Mangifera Mangifera Ding Hou M. laurina Polyembryonic Indonesia Wild distribution: Myanmar, Vietnam, Malesia, Thailand. Cultivated in Borneo, Sumatra, Java, (Indonesia), and the Philippines. 40 Juicy, very acid and fibrous fruit: edible.
Resistance to anthracnose (fruit skin).
Used as a rootstock for M. indica in Malaysia.Limus Deciduae M. pajang Monoembryonic Indonesia Endemic to Borneo (Brunei Indonesia, Malaysia).
Common in cultivation.Unknown Fruit flesh: deep-orange-yellow, fibrous, acid to acid sweet, mildly fragrant: edible.
Largest fruit in the genus.Mangifera Euantherae Pierre M. caloneura (Xoài Quéo) Polyembryonic Vietnam Dry deciduous dipterocarp forests in Myanmar, Thailand, Laos, and Vietnam 40 Fruit with good aroma, sour taste: edible.
Drought tolerance / resistance to anthracnose (fruit skin) and gummosis.Limus Perrennis M. odorata Polyembryonic Malaysia Never been observed in the wild.
Origin is unknown, primarily cultivated in Guam, Philippines, Thailand, and Vietnam.
Introduced to Indonesia, Malaysia, and Singapore.40 Firm, fibrous, sweet to acid-sweet, juicy fruit with a strong smell and turpentine flavour: edible.
Resistance to anthracnose.Limus Perrennis M. foetida Monoembryonic Malesia Western part of Malesia (Sumatra, Java, Borneo, Malay Peninsula), wild and cultivates, introduced to Burma, Indochina. 40 Fruit with strong turpentine smell, sweet and pleasant, very fibrous: edible. Mangifera Mangifera Ding Hou M.altissima Polyembryonic Philippines Native to Philippines, Indonesia, Papua New Guinea, Solomon Islands. Unknown Smooth fibrous or non-fibrous fruit: edible. Limus Deciduae M. caesia Monoembryonic Indonesia Natural distribution: Peninsular Malaysia, Borneo (Brunei Darussalam, Indonesia, Malaysia) and Sumatra (Indonesia).
Cultivated from Peninsular Thailand, Bali, Java and to the Philippines.40 Sweet flesh with strong fragrance in the fruit: edible. Mangifera Mangifera Ding Hou M. zeylanica Monoembryonic Sri Lanka Endemic to Sri Lanka, not found under cultivation. Unknown Very juicy fruit, fibrous, pleasant, sweet taste pulp: edible. Mangifera Mangifera Ding Hou M. sylvatica Polyembryonic Myanmar Native to India (Sikkim), China, Myanmar, Thailand, Bangladesh, and Nepal. 40 Fruits almost fibreless, little pulp, sweet and sour taste: edible. Mangifera Mangifera Ding Hou M. quadrifida Unknown Malesia Sumatra, Malay Peninsula, Borneo, and Sunda Islands. Unknown Fruits with less fibres, sweet and pleasant smell. Mangifera Mangifera Ding Hou M. lalijiwa Polyembryonic Indonesia Indonesia (Java, Madura, Bali and probably in Sumatra). Very rare both in wild and in cultivation. Unknown Small glossy yellowish fruit with acid-sweet taste: edible. Mangifera Mangifera Ding Hou M. applanata Polyembryonic Malesia Native to Indonesia (Kalimantan and Sumatra) and Malaysia (Pahang, Sabah, and Sarawak).
Cultivated in some areas of Borneo.Unknown Juicy, very acid fruit with turpentine and lemon taste: edible. Mangifera Mangifera Ding Hou M. casturi Polyembryonic Indonesia Endemic to South Kalimantan, Indonesia.
Not known in the wild, only found in cultivation.Unknown Small fruits, with an attractive purple color and a distinctive aroma: edible. Mangifera Mangifera Ding Hou M. indica cv. 'Kensington Pride' Polyembryonic Australia First planted in Bowen, North Queensland, Australia.
Pre-Australian origin is unknown.
Grown throughout Australia.40 Soft and juicy flesh with moderate to little
fibre, sweet with a characteristic flavour excellent eating quality.Mangifera Mangifera Ding Hou M. i ndica cv. 'Alphonso' Monoembryonic India Prominently grown in India. 40 Firm to soft flesh, low in fibre content, sweet with characteristic aroma, very pleasant taste: edible. Mangifera Mangifera Ding Hou M. i ndica cv. 'Tommy Atkins' Monoembryonic USA (Florida) Originated and most widely grown commercial cultivar in USA. 40 Fruit with firm, medium juicy, medium amount of fibre of good eating quality.
Highly resistant to anthracnose disease.Table 1.
Details of the 14 Mangifera species used in this study including country of origin, native distribution and important characteristics.
-
Species/
genotypeGenome
size (bp)Overall GC content LSC (bp) SSC (bp) IR (bp) Total no.
of genesTotal no.
of protein coding genesTotal no.
of tRNAsTotal no.
of rRNAsM. laurina 158,965 37.8% 87,714 18,427 26,412 115 80 31 4 M. laurina † 158,965 37.8% 87,714 18,427 26,412 115 80 31 4 M. pajang 158,830 37.8% 87,654 18,424 26,376 115 80 31 4 M. pajang ‡ 158,887 37.8% 87,709 18,426 26,376 115 80 31 4 M. caloneura 157,780 37.9% 86,672 18,350 26,379 115 80 31 4 M. odorata 158,889 37.8% 87,708 18,427 26,377 115 80 31 4 M. foetida 158,882 37.8% 87,706 18,422 26,377 115 80 31 4 M. altissima 157,780 37.9% 86,673 18,349 26,379 115 80 31 4 M. altissima † 157,780 37.9% 86,673 18,349 26,379 115 80 31 4 M. caesia 151,752 37.6% 98,334 19,064 17,177 115 80 31 4 M. zeylanica 157,604 37.9% 86,507 18,319 26,389 115 80 31 4 M. lalijiwa 157,779 37.9% 86,672 18,349 26,379 115 80 31 4 M. applanata 157,779 37.9% 86,672 18,349 26,379 115 80 31 4 M. casturi 158,942 37.8% 87,733 18,425 26,392 115 80 31 4 M. quadrifida † 158,889 37.8% 87,679 18,424 26,393 115 80 31 4 M. sylvatica † 158,025 37.9% 86,856 18,387 26,391 115 80 31 4 M. sylvatica* 157,781 37.9% 86,712 18,347 26,361 115 80 31 4 M. odorata* 158,883 37.8% 87,702 18,427 26,377 115 80 31 4 M. persiciformis* 158,952 37.8% 87,566 18,536 26,368 115 80 31 4 M. hiemalis* 158,838 37.8% 87,681 18,535 26,368 115 80 31 4 M. i ndica cv. 'Kensington Pride' 157,780 37.9% 86,673 18,349 26,379 115 80 31 4 M. indica cv. 'Alphonso' 157,780 37.9% 86,673 18,349 26,379 115 80 31 4 M. indica cv. 'Tommy Atkins'* 157,780 37.9% 86,673 18,349 26,379 115 80 31 4 GC, guanine or cytosine; IR, inverted repeats; LSC, large single copy; SSC, small single copy. * Mangifera species for which chloroplast genomes were assembled using raw data downloaded from NCBI. † Mangifera species collected from the Botanical Ark, Queensland, Australia. ‡ Mangifera species collected from the Durian Heavan Farm, Queensland, Australia. M. pajang is collected from TreeFarm, Queensland, Australia. Table 2.
Annotation of the chloroplast genomes of Mangifera species.
-
Clade name in the phylogenetic tree Species/
genotypes in comparisonSpecies/
genotype ASpecies/
genotype BTotal no. of
variations between A vs BTypes of variation Insertions Deletions SNPs Substitutions BI M. laurina
M. laurina †
M. casturi
M. quadrifida †M. laurina M. laurina † 0 − − − − M. laurina M. casturi 116 30 30 51 5 M. casturi M. quadrifida 191 32 25 119 15 BII M.odorata
M. odorata*
M. pajang
M. pajang ‡
M. foetida †M. odorata M. odorata* 2 − 2 − − M. pajang M. pajang ‡ 27 7 8 11 1 M. odorata M. pajang 75 17 16 39 3 M. odorata M. foetida 108 21 16 68 3 BIII M. persiciformis*
M. hiemalis*M. persiciformis* M. hiemalis* 45 9 9 27 − CI M. altissima
M. altissima †
M. lalijiwa
M. applanata
M. caloneura
M. indica cv. 'Kensington Pride'
M. indica cv. 'Tommy Atkins'
M. indica cv. 'Alphonso'M. indica cv.
'Tommy Atkins'M. indica cv.
'Kensington Pride'0 − − − − M. indica cv.
'Tommy Atkins'M. indica cv.
'Tommy Atkins'*0 − − − − M. indica cv. 'Alphonso' M. indica cv. 'Kensington Pride' 0 − − − − M. indica cv. 'Alphonso' M. indica cv.
'Tommy Atkins'0 − − − − M. altissima M. altissima † 0 − − − − M. altissima M. indica cv. 'Kensington Pride' 0 − − − − M. lalijiwa M. indica cv. 'Kensington Pride' 1 − 1 − − M. applanata M. indica cv. 'Kensington Pride' 1 − 1 − − M. caloneura M. indica cv. 'Kensington Pride' 2 1 1 M. lalijiwa M. applanata 0 − − − − M. caloneura (Xoài Quéo) M. lalijiwa 3 2 1 − − CII M. sylvatica*
M. indica cv. 'Kensington Pride'M. sylvatica* M. indica cv. 'Kensington Pride' 96 22 14 57 3 CIII M. zeylanica
M. indica cv. 'Kensington Pride'M. zeylanica M. indica cv. 'Kensington Pride' 175 18 26 122 9 * Species for which chloroplast genomes were assembled using raw data downloaded from NCBI. † Mangifera species collected from the Botanical Ark, Queensland, Australia. ‡ Mangifera species collected from the Durian Heavan Farm, Queensland, Australia. Table 3.
INDELs, SNPs and substitutions identified with respect to clustering pattern in chloroplast phylogeny.
-
Gene name Gene
tree no.Species proposed to be hybrid Species supposed to be undergone domestication/hybridization Trees in which M. sylvatica clusters with M. persiciformis
in the same clade as sister taxaM. odorata M. casturi M. zeylanica M. sylvatica Trees in which M. odorata clusters with M. indica in the same clade as sister taxa Trees in which M. odorata clusters with M. foetida in the same clade as sister taxa Trees in which M. casturi clusters with M. indica in the same clade as sister taxa Trees in which M. casturi clusters with M. quadrifida in the same clade as sister taxa Trees in which M. zeylanica clusters with M. indica in the same clade as sister taxa Trees in which M. sylvatica clusters with M. hiemalis in the same clade as sister taxa 4-alpha-glucanotransferase 1 × × × × √ × × A49-like RNA polymerase I associated factor 2 × × × × √ × × Acyl-CoA dehydrogenase,
C-terminal domain3 × × × × × √ × Alphabeta hydrolase family 4 √ × × × √ × √ Aminomethyltransferase folate-binding domain 5 × × × × × × × Aminopeptidase I zinc metalloprotease (M18) 6 × × × × × × × ArgJ family 7 × × × × × × × Armadillobeta-catenin-like repeat 8 × × × × × × × Brix domain 9 × × × × × × √ Cactus-binding C-terminus of cactin protein 10 × × × × × × × Carbon-nitrogen hydrolase 11 √ × × × × × × CobWHypBUreG, nucleotide-binding domain 12 × × × × × × × Cohesin loading factor 13 × × × × × × × Creatinase Prolidase N-terminal domain 14 × × × × × × × Cyclophilin type peptidyl-prolyl cis-trans isomeraseCLD 15 × × × × × × × Cytochrome b5-like HemeSteroid binding domain 16 × × × × × × × DDRGK domain 17 × √ × × × × × Dienelactone hydrolase family 18 × √ × × × √ × Divergent CRALTRIO domain 19 × × × × × √ × Dual specificity phosphatase, catalytic domain 20 × × × × √ √ × Dynamin family (2) 21 × × × × × × × ER membrane protein complex subunit 1, C-terminal 22 × × × × × × × Eukaryotic protein of unknown function (DUF866) 23 × × √ × × × × FAD binding domain 24 × × × × √ × × Glucose-6-phosphate isomerase 25 × × × × × × √ Glucosidase II beta subunit-like protein 26 × × × × × √ × Glycosyl hydrolases family 31 27 × × × × × √ × Glyoxalase bleomycin resistance protein dioxygenase superfamily 28 × × × × × × × Hydroxyacylglutathione hydrolase C-terminus 29 × × × × × × × Methyltransferase domain 30 × √ × × × × × NmrA-like family 31 × × × × √ √ × N-terminal domain of lipoyl synthase of Radical_SAM family 32 × × × × × × × PAP2 superfamily C-terminal 33 × × × × × × × Prolyl oligopeptidase, N-terminal beta-propeller domain 34 × × × × × × × Putative tRNA binding domain 35 × × × × √ √ × Pyruvate phosphate dikinase, AMPATP-binding domain 36 × × × × × √ √ Redoxin 37 √ × × × × × × Ribosomal protein L13 38 × × × √ √ √ × RNA recognition motif.
(a.k.a. RRM, RBD, or RNP domain)39 × × × × × √ × Rubrerythrin 40 × × × × × × √ snoRNA binding domain, fibrillarin 41 × × × × √ × × Sodium Bile acid symporter family 42 × √ × × √ × × TFIIS helical bundle-like domain 43 × × × × √ × × Transcription factor TFIIB repeat 44 √ × × × × √ √ tRNA synthetase class II core domain (G, H, P, S and T) 45 × × × × × × √ Uncharacterized ACR, YdiUUPF0061 family 46 × × √ × × × √ WLM domain 47 × × × × × × √ Table 4.
Gene trees indicating clustering of suggested hybrids/ wild relatives with their proposed parents as sister taxa.
Figures
(3)
Tables
(4)