-
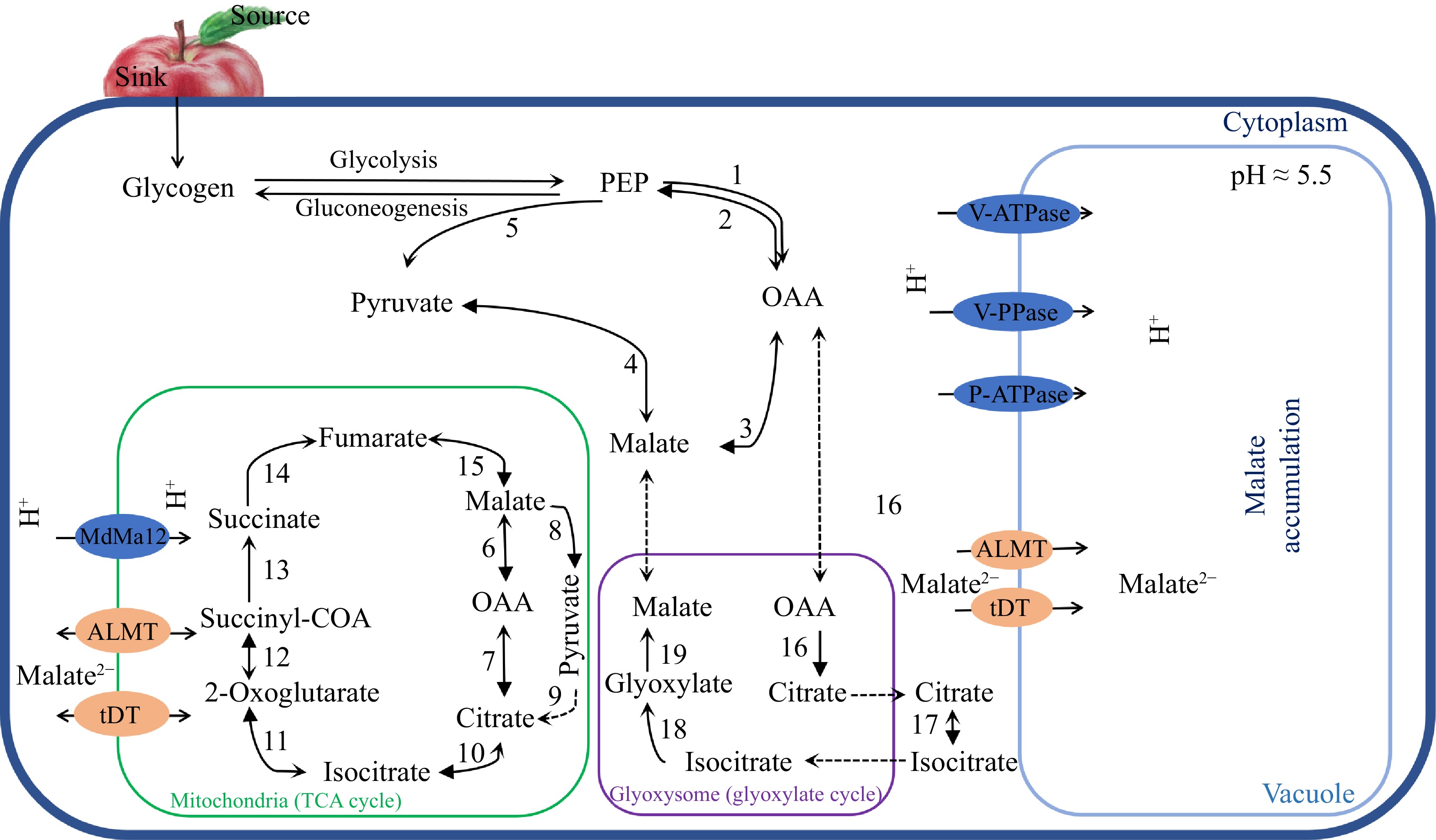
Figure 1.
A simplified model for malate metabolism, transport and accumulation in fruit. Major enzymes involved in malate metabolism in fruit: 1, phosphoenolpyruvate carboxylase (PEPC); 2, phosphoenolpyruvate carboxykinase (PEPCK); 3, NAD-cyMDH (NAD-cytoplasmic malate dehydrogenase); 4, NADP-cytoplasmic malate enzyme (NADP-cyME); 5, pyruvate kinase (PK); 6, NAD-mitochondrial malate dehydrogenase (NAD-mtMDH); 7, mitochondrial citrate synthase (mtCS); 8, NAD-mitochondrial malate enzyme (NAD-mtME); 9, pyruvate dehydrogenase (PDH); 10, mitochondrial aconitase (mtACO); 11, isocitrate dehydrogenase (ICDH); 12, α-oxoglutarate dehydrogenase (α-OGDH); 13, succinyl-coa synthase (SCS); 14, succinate dehydrogenase (SuDH); 15, fumarase; 16, cytoplasmic citrate synthase (cyCS); 17, glyoxylate aconitase (glyACO); 18, isocitrate lyase (ICL); 19, malate synthase (MS).
-
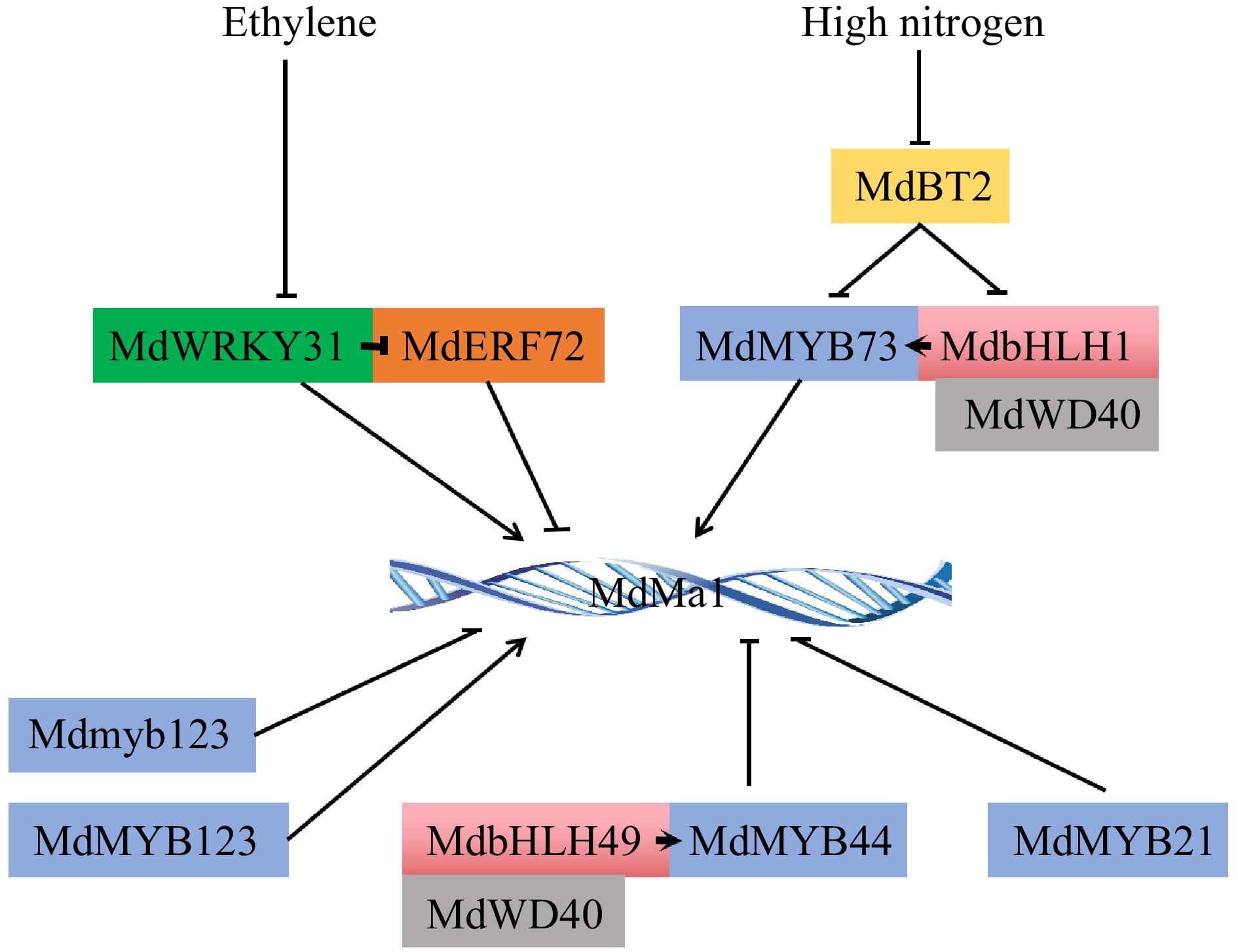
Figure 2.
Upstream regulators of major gene Ma1 regulating apple fruit acidity. The arrows represent positive regulation and the rest represent negative regulation.
-
Gene family Gene name Protein name Activation (+)/
inhibition (−)Module control Species MYB transcription factor MdMa1 MdMYB73 + MdBT2-MdCIbHLH1
-MdMYB73Malus domestica MdMa1/MdMa11 MdMYB123 + — Malus domestica MdMa1 MdMYB44 − MdbHLH49-MdMYB44 Malus domestica MdMa1 MdMYB21 − — Malus domestica MdVHA-A3/D2 Ma10 MdMYB44 − WD40-MdbHLH49
-MdMYB44Malus domestica MdVHA-B1/E MdVHP1
MdtDTMYB1/10 + MdTTG1-MdbHLH3
-MdMYB1/10Malus domestica MdVHA-A MdVHP1 MdMYB73 + MdBT2-
WD40-MdbHLH1
-MdMYB73Malus domestica CitPH5 CitPH4 + CitTRL-CitPH4 Citrus reticulata WRKY transcription factor SlALMT9 SlWRKY42 − — Solanum lycopersicum PpALMT9 PpWRKY44 + PpABF3-PpWRKY44 Pyrus spp. ZjALMT4 ZjWRKY7 + — Ziziphus jujuba MdMa1 MdWRKY31 + — Malus domestica MdMDH1 MdWRKY126 + — Malus domestica bHLH transcription factor MdMDH1 MdbHLH3 + — Malus domestica NAC transcription factor AcALMT1 AcNAC1 + — Actinidia spp. ERF transcription factor MdMa1 MdERF72 − MdWRKY31-MdERF72 Malus domestica CitVHA-C4 CiERF13 + — Citrus reticulata PP2C family MdVHA-A3/B2/D2 Ma10 MdPP2CH − SAUR37-MdPP2CH Malus domestica Table 1.
The crucial genes and their upstream regulatory factors of fruit acidity regulation.
Figures
(2)
Tables
(1)