-
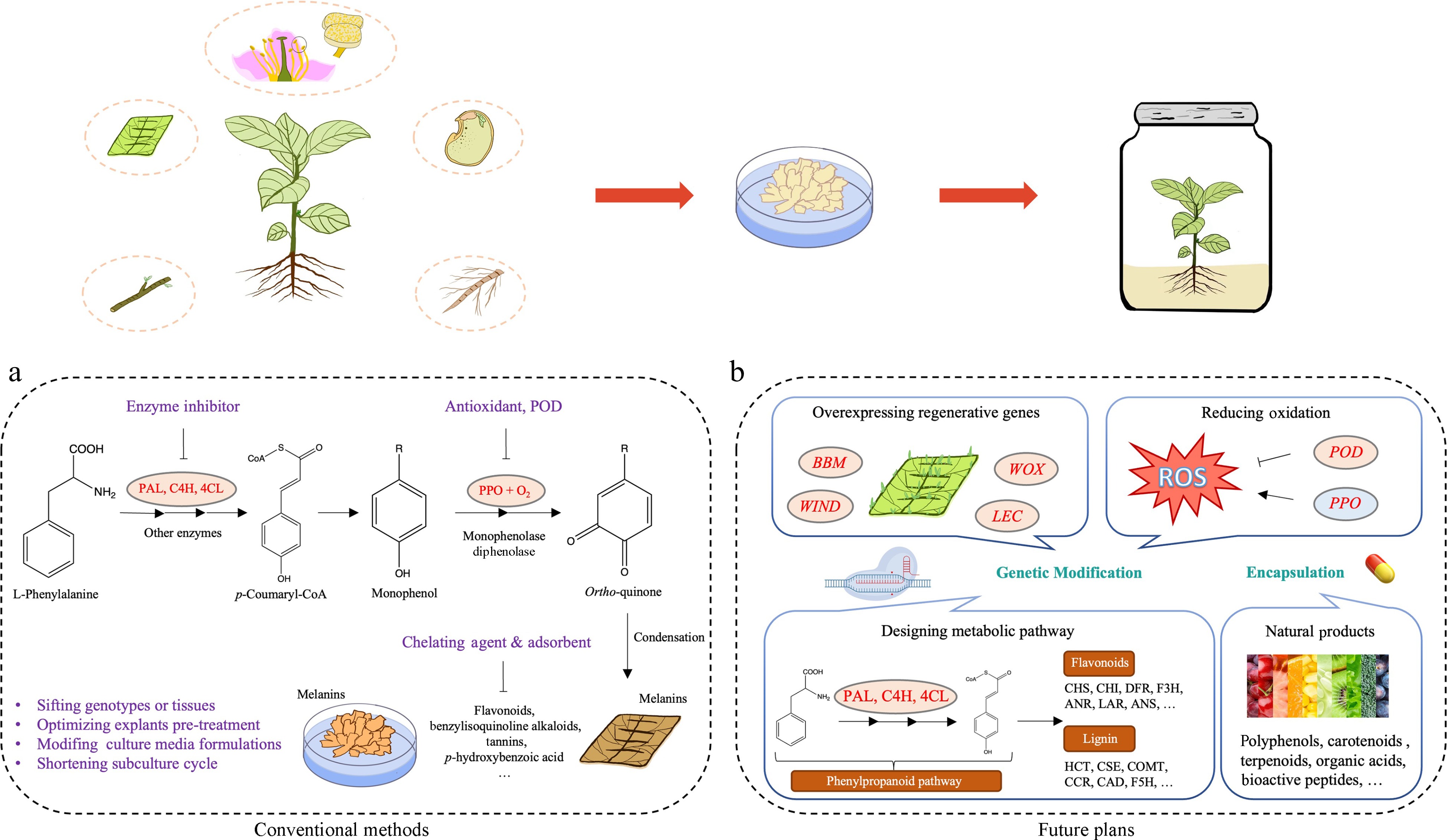
Figure 1.
Techniques and perspectives on controlling browning in in vitro cultures of woody plants. (a) Traditional management strategies for controlling enzymatic browning. (b) Prospective strategies for addressing browning: genetic approaches by targeting key metabolic pathways and regenerative genes, encapsulation of natural products.
-

Figure 2.
Comparison of healthy and browning callus in Carya cathayensis Sarg. Microscopic view of (a) healthy cells and (b) browning cells. Scale bars = 50 μm. Phenotypic progression of callus after subculture: (c) non-browning callus at day 10, (d) slightly browning callus at day 17, and (e) browning callus at day 24. Scale bars = 4 mm. FDA staining indicating viability of (f) non-browning callus, (g) slightly browning callus, and (h) browning callus. Scale bars = 4 mm. (i)−(j) Total phenolic content and total flavonoid content of non-browning callus at day 10, slightly browning callus at day 17, and browning callus at day 24. Error bars represent the SD (n = 3). Statistical analysis was performed using one-way ANOVA for each treatment (** p < 0.01, *** p < 0.001, **** p < 0.0001).
-
Pathway Gene Species Function Ref. Oxidative reaction LAC L. chinensis Overexpression LcLAC promotes callus browning and is involved in polyphenols polymerization. [7] LAC7 Malus domestica MdWRKY31 binds the promoter of MdLAC7, positively regulating its activity to promote peel browning. [58] POD E. urophylla PUB and 6-BA enhance POD activity, alleviating browning. [49] PPO2 Juglans regia JrPPO2 shows high activity in browning calluses. [51] Phenylpropanoid pathway DFRa Camellia sinensis CsDFRa is involved in the regulation of metabolic flux affecting secondary metabolism and phenotypic characteristics. [59] FLS, UGT Ginkgo biloba L. Expression of FLS and UGT in the flavonoid pathway are significantly higher in browning callus than in green callus. [60] LAR1 M. domestica MdLAR1 inhibits the expression of other genes in the anthocyanin biosynthesis. [61] MYB21, MYB54 Pyrus ussuriensis
Maxim.PuMYB21/PuMYB54 enhance the degradation of membrane phospholipids leading to pericarp browning. [62] PAL, 4CL Pyrus spp Down-regulated expression of PuPAL and Pu4CL result in browning. [63] PAL, 4CL, F3H, CYP73A, CHS, CHI, ANS, DFR, PGT1 Malus sieversii Genes related to flavonoid biosynthesis increase flavonoid accumulation during browning. [64] PAL, C4H, 4CL,
CCR, CADM. domestica Genes increase the biosynthesis of phenolic compounds contributing to browning. [65] PAL, ANS J. regia These genes are involved in the synthesis of phenolic compounds and color changes in walnuts. [66] Table 1.
Genes related to browning in economically important woody plants.
-
Gene Species Organ Function Ref. ATXR2 A. thaliana Callus, root ATXR2 promotes callus formation and lateral root growth [99] BBM Saccharum officinarum L. Callus BBM exhibits high levels of expression in embryogenic callus [100] CKI Gossypium hirsutum Hypocotyl section Overexpression of GhCKI inhibits somatic embryo formation and plant regeneration [101] GRF4-GIF1 Triticum aestivum GRF4-GIF1 improves regeneration efficiency and overcomes genotypic limitation [102] IAA Moringa oleifera Lam. Shoot IAA13 inhibits shoot regeneration [103] LBD5 Z. mays Overexpression of ZmLBD5 in Arabidopsis reduces ROS [104] LBD19 A. thaliana Callus Negatively regulates callus formation [105] LEC S. officinarum L. Callus LEC exhibits high levels of expression in embryogenic callus [100] PSK C. lanceolata Somatic embryogenesis Overexpression of ClPSK in Arabidopsis enhances somatic embryogenesis capabilities and lowers hydrogen peroxide levels [92] REF1 Solanum lycopersicum L. Callus, shoot Upon cellular damage, REF1 serves as an initial wound signal molecule and is recognized by the receptor PORK1, activating the expression of key cell reprogramming regulator SlWIND1 [94] WIND A. thaliana Callus, shoot WIND1 promotes callus formation and shoot regeneration [106] WOX Salix suchowensis Shoot WOX promotes shoot regeneration [107] WOX2 P. abies Somatic embryos Down-regulation of PaWOX2 early in embryo development significantly decreases in the yield of mature embryos [97] P. pinaster Callus WOX2 serves as a marker gene for somatic embryogenesis [96] WOX5 T. aestivum Callus WOX5 increases transformation efficiency with less genotype dependency [108] C. lanceolata Leaf Overexpression of WOX5 improves shoot regeneration but causes aborted embryo development, resulting in a partially sterile phenotype [98] WOX6 C. lanceolata Leaf Overexpression of WOX6 improves shoot regeneration [98] WOX11 '84K' (P. alba ×
P. glandulosa)Leaf PtWOX11 promotes de novo root, shoot organogenesis in poplar [109] WOX13-1 Malus × domestica Callus MdWOX13-1 increases callus weight and enhanced ROS scavenging ability [110] Table 2.
Regeneration-promoting genes used in plant genetic transformation.
Figures
(2)
Tables
(2)