-
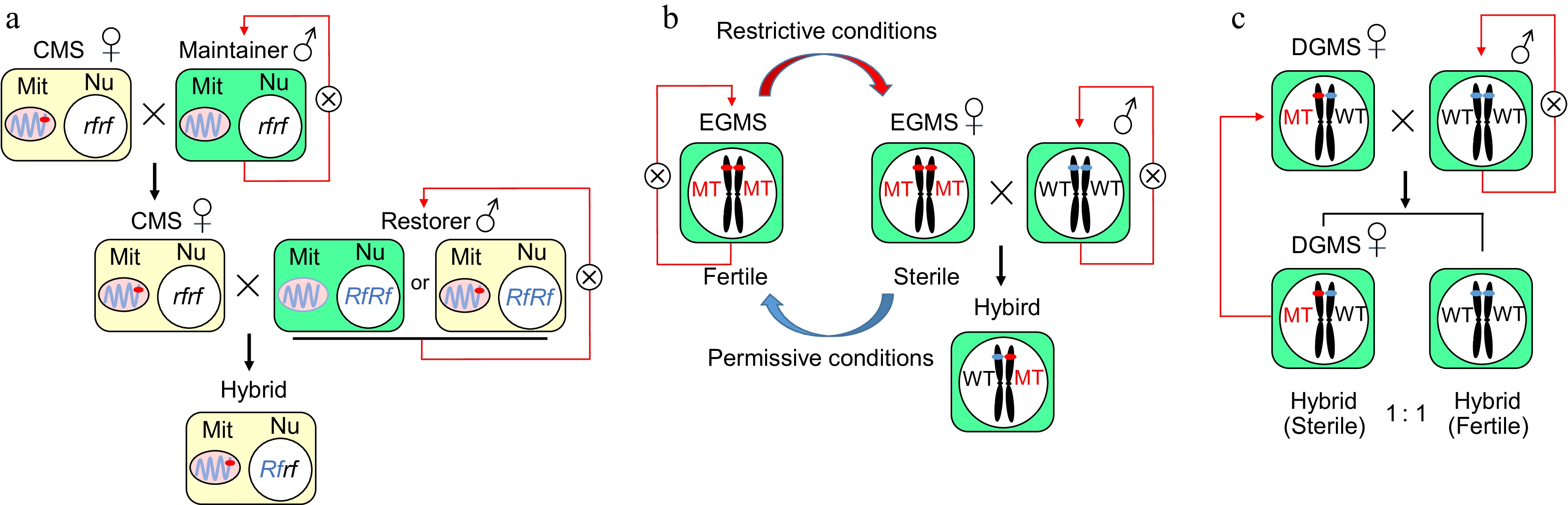
Figure 1.
The genetic and mechanistic models of cytoplasmic male sterility and genic male sterility. (a) A CMS line contains a sterile cytoplasm and a nonfunctional restorer of the fertility gene (rf), which can be maintained by recurrent crossing to a maintainer line with normal cytoplasm. A restorer line with normal or sterile cytoplasm and a functional restorer gene (Rf) can restore fertility in the F1 hybrid. Mit: Mitochondrion, Nu: Nucleus. (b) The application and molecular mechanism of genes related to environmental male sterility in a two-line hybrid system in rice. The fertility of the EGMS line could switch based on the state of the environment to produce hybrid seeds in a two-line system. The EGMS line acts as the maternal parent to produce F1 hybrid seeds when it is sterile under restrictive conditions and does self-breeding when it is fertile under permissive conditions. (c) A DGMS line was regulated by a single gene/locus and exhibited male sterility under heterozygous conditions. When crossed with fertile plants, the progeny had a 1:1 ratio of sterile to fertile individuals, and the sterile plants could be repeatedly used as DGMS lines.
-
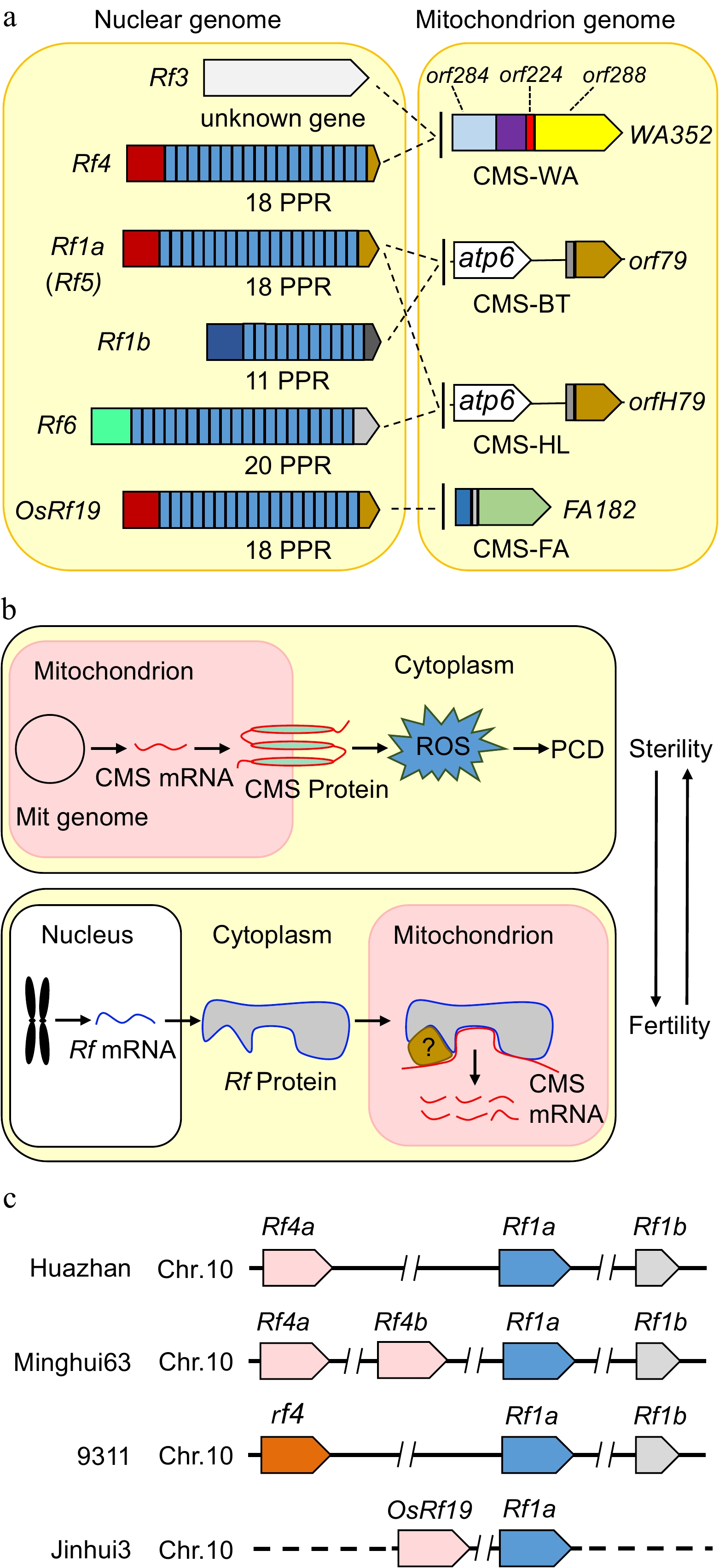
Figure 2.
The CMS genes, restorer genes and mechanistic models of cytoplasmic male sterility in rice. (a) The mitochondrial genes WA352, orf79, orfH79 and FA182 are expressed in CMS lines, damaging the development of anthers and pollen, and are suppressed by the corresponding nuclear Rf genes in hybrids, thereby restoring male fertility. Most Rf genes encode a PPR protein. Rf4, Rf1a, Rf1b, Rf6 and OsRf19 contain 18, 18, 11, 20, and 18 PPR domains, respectively. (b) Proposed model illustrating the mechanism of CMS and fertility restoration in rice. CMS genes derived from the mitochondrial genome are transcribed and translated into proteins with transmembrane domains in mitochondria, leading to an explosion of reactive oxygen species (ROS) and ultimately premature programmed cell death (PCD), resulting in sterile male gametes in CMS lines. Restorer genes derived from the nuclear genome encode proteins with mitochondrial transpeptides in the F1 generation. The restorer proteins translocate into mitochondria and bind to CMS transcripts directly or indirectly to cleave or degrade RNA; alternatively, the interactions between restorer proteins and RNA block the translation of CMS proteins. Therefore, CMS products are eliminated, and male gametes become fertile. (c) The relative positions of the restorer genes Rf4, Rf1a, Rf1b, and OsRf19 on chromosome 10 in different rice varieties. The dashed line represents the unknown sequence in Jinhui3.
-
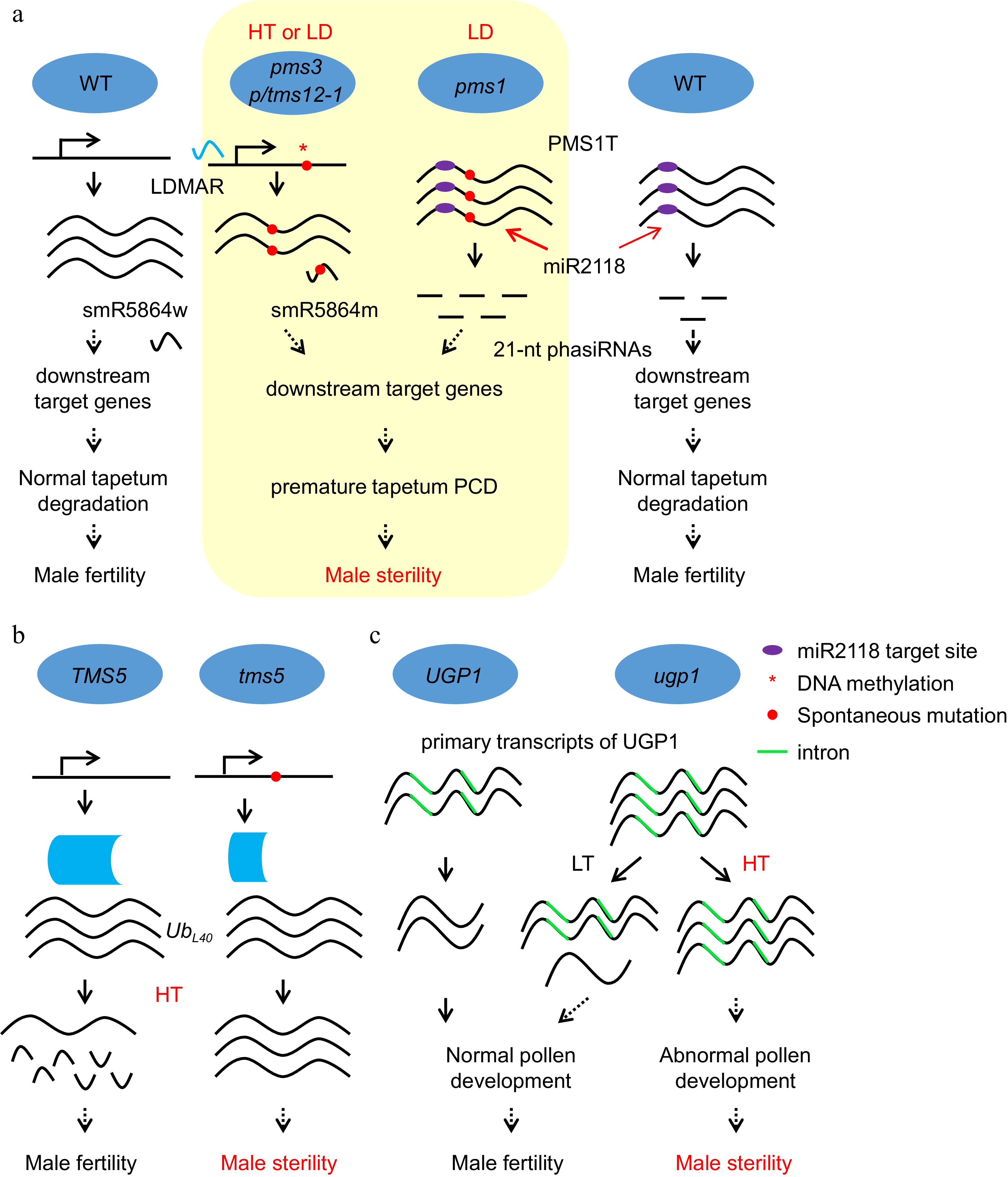
Figure 3.
Noncoding RNAs and RNA processing play important roles in EGMS. (a) The molecular mechanism of photoperiod-sensitive male sterility regulated by noncoding RNAs produced from the pms3/ptms12-1 and pms1 loci. HT, high temperature; LD, long day. (b) The molecular mechanism of thermosensitive male sterility regulation through RNA processing in tms5 and ugp1. The mutation in tms5 leads to a short version of the tms5 protein, which fails to process the mRNA of UbL40 under HT and results in male sterility. (c) The molecular mechanism of thermosensitive male sterility regulation through RNA processing of ugp1. More primary transcripts containing introns accumulated in ugp1 than in WT, and the introns could not be spliced under HT, which caused no UGP1 protein to maintain normal pollen development in ugp1, resulting in male sterility under HT. LT, low temperature.
-
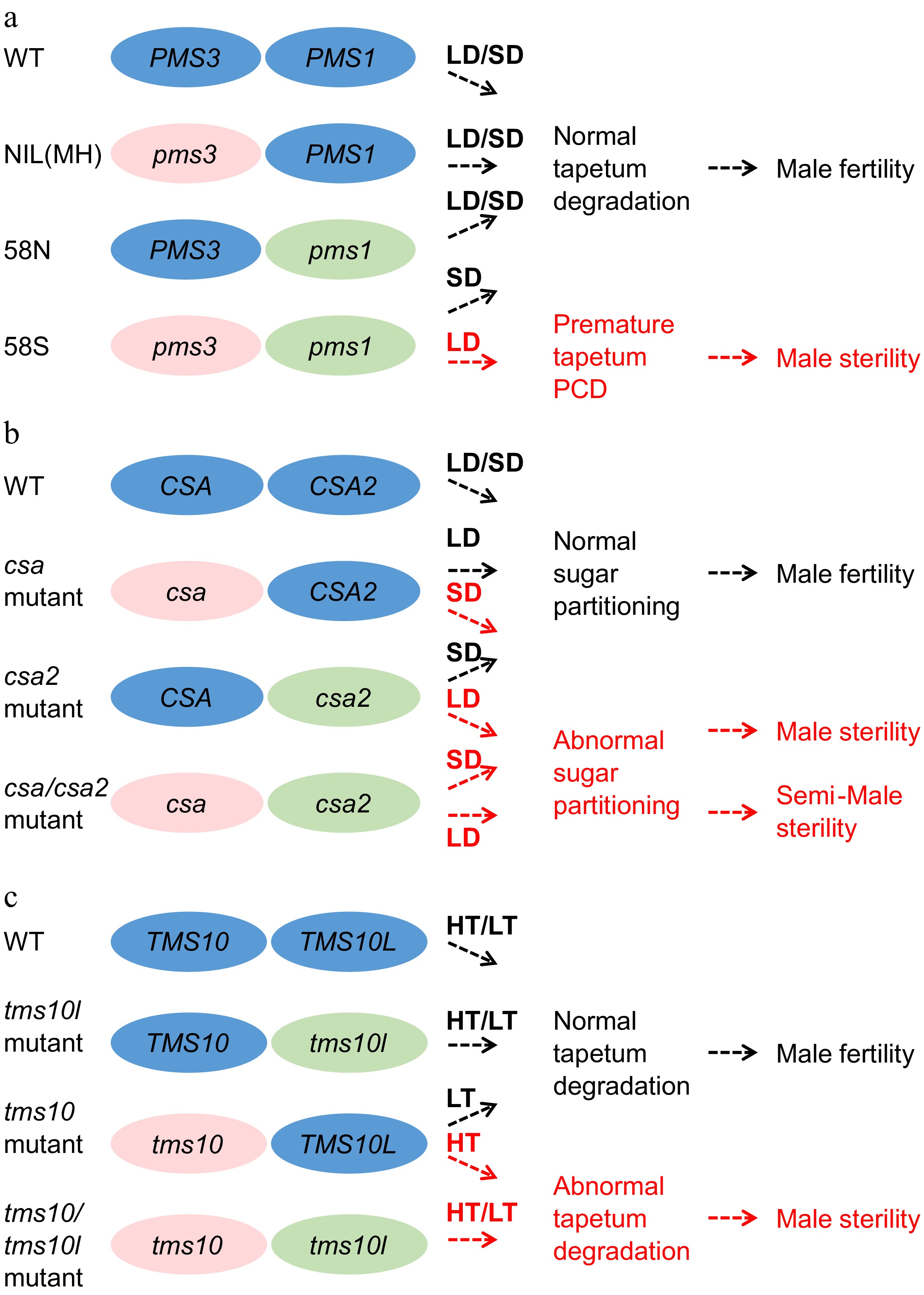
Figure 4.
The gene pairs involved in EGMS. Three gene pairs conferring environmentally sensitive male sterility, namely, pms1 and pms3, csa and csa2, and tms10 and tms10l, are listed in the figure. LD, long-day conditions. SD, short-day conditions. LT, low-temperature conditions. HT, high-temperature conditions. (a) Both pms1 and pms3 are required to induce the PTGS trait in 58S. Rice plants lacking either pms3 or pms1 are fertile under both LD and SD. NIL(MH), a near-iso-genic line of pms1 in the background of 58S in which the PMS1 gene was introduced from MH63, was constructed by backcrossing with 58S. (b) The rPTGS csa mutant shows a sterile phenotype under SD, while the PTGS mutant csa2 is semisterile under LD. The double mutant csa/csa2 was sterile under LD and semisterile under SD. (c) tms10 is TGMS and sterile under HT, whereas tms10l is fertile under both HT and LT. The double mutant tms10/tms10l is sterile under both HT and LT.
-
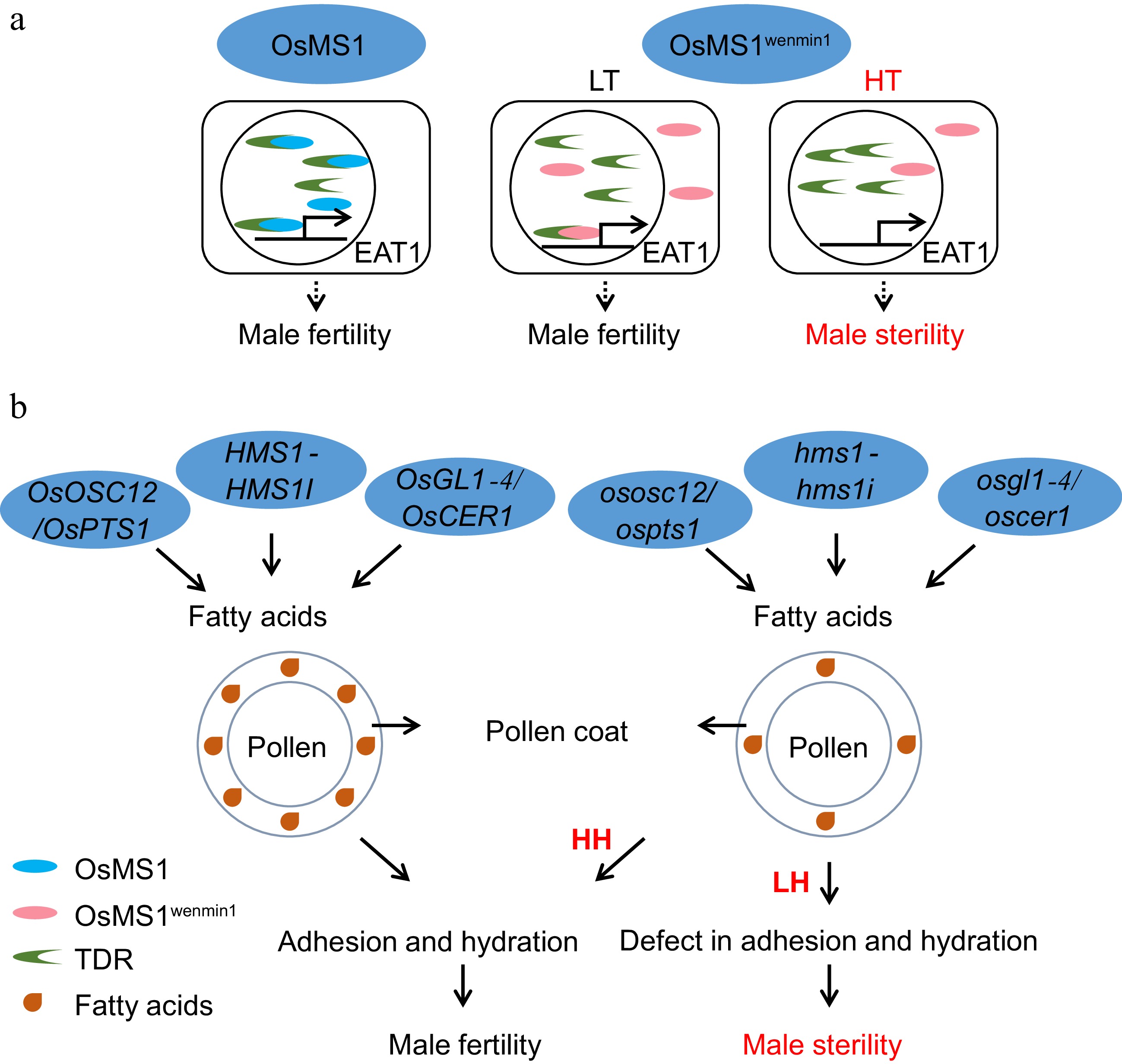
Figure 5.
Pollen development in relation to EGMS. (a) The molecular pathway of TGMS regulated by OsMS1wenmin1. LT, low-temperature conditions. HT, high-temperature conditions. (b) The mechanisms by which humidity-sensitive male sterility is regulated by OsOSC12/OsPTS1, OsGL1-4/OsCER1, and HMS1-HMS1I. LH, low-humidity condition. HH, high-humidity condition. Due to the reduction in fatty acids in the pollen coat in the HMS line under low humidity, the HMS line fails to finish adhesion and hydration, leading to sterility. However, under high humidity, a defective pollen coat is sufficient to complete adhesion and hydration in the HMS line to maintain male fertility.
-
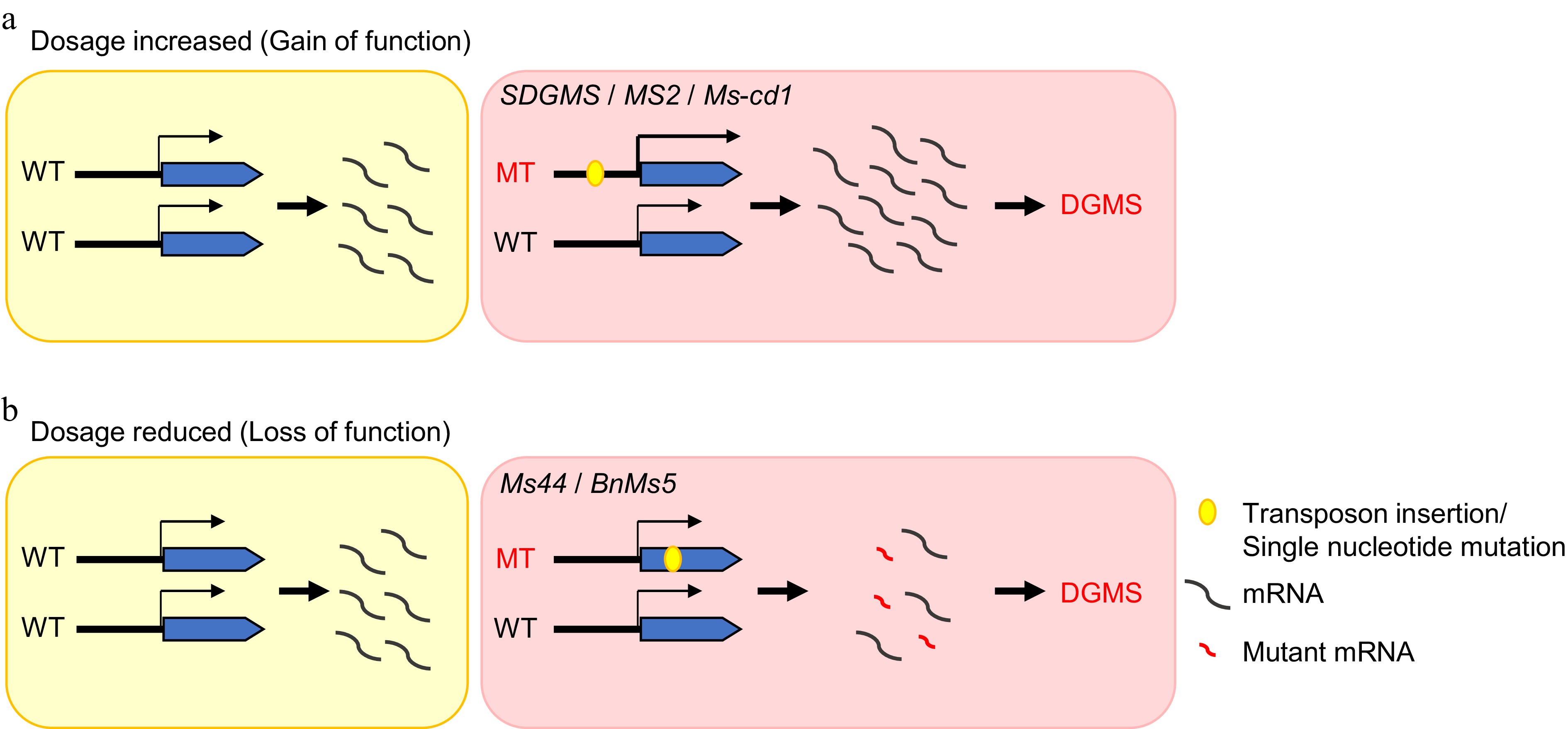
Figure 6.
The mechanistic models of dominant genic male sterility in crops. (a) Gain-of-function caused by a mutation in the promoter region, which increases the expression level of genes. (b) Loss of function due to a mutation in the coding region, which results in protein dysfunction.
-
CMS type Cytoplasm source CMS gene Rf gene Location Ref. CMS-BT O. sativa ssp. indica (Chinsurah Boro II) orf79 Rf1a Chr.10 [6] Rf1b Chr.10 [7] CMS-CW O. rufipogon (W1) CW-orf307 Rf17 Chr.4 [31] CMS-D1 O. rufipogon (Dongxiang wild rice) orf182 UK UK [15] CMS-FA O. rufipogon (Fujian wild abortive) FA182 OsRf19 Chr.10 [12] CMS-HL O. rufipogon (Hong Lian wild rice) orfH79 Rf5 Chr.10 [8,26] Rf6 Chr.8 [29] CMS-LD O. sativa ssp. indica (Lead rice) L-orf79 Rf2 Chr.2 [9,10] CMS-RT98 O. rufipogon (W1109) orf113 Rf98 Chr.10 [37] CMS-RT102 O. rufipogon (W1125) orf352 UK UK [5] CMS-TA O. sativa ssp. indica (Tadukan) orf312 UK UK [20,21] CMS-WA O. rufipogon (Wild abortive) WA352 Rf3 Chr.1 [4] Rf4 Chr.10 [28] Rf20 Chr.1 [30] UK: unknown gene/location. Table 1.
Types and characteristics of CMS/Rf systems in rice.
-
EGMS line EGMS type Locus Encoding product Biological pythay Ref. NK58S PGMS pms3 Long noncoding RNALDMAR Noncoding RNA and RNA processing [49,50] NK58S PGMS pms1 Long noncoding RNA PMS1T Noncoding RNA and RNA processing [51] csa mutant rPGMS csa R2R3 MYB transcription factor Sugar distribution [52−54] csa2 mutant PGMS csa2 R2R3 MYB transcription factor Sugar distribution [52] AnnongS-1 TGMS tms5 RNase Zs1, 2’3’-cyclic phosphatase of tRNA RNA processing [55,78] ugp1mutant TGMS ugp1 UDP-Glucose Pyrophosphorylase1 RNA processing [56] tms10 mutant TGMS tms10 A leucine-rich receptor kinase LRR-RLK Signaling transduction [57] Hengnong S-1, Tian1S TGMS tms9-1/OsMS1wenmin1 PHD zinc finger protein Pollen developent [61,85,86] Peiai64S TGMS p/tms12-1 Small RNA Noncoding RNA [69] ostms18 mutant TGMS ostms18 GMC oxidoreductase Pollen wall formation [59] ostms15 mutant TGMS ostms15 A leucine-rich receptor kinase LRR-RLK Pollen developent [58] ostms19 mutant TGMS ostms19 PPR protein ROS accumulation [60] E157, S1708 and S4928 HGMS ososc12/ospts1 Bicyclic triterpenoid polyacetylacetone synthase Pollen coat [62] hms1 mutant HGMS hms1 3-β-ketoacyl-CoA synthase 6 Pollen coat [64] hmsli mutant HGMS hmsli Very-long-chain enoyl-CoA reductase Pollen coat [64] osgl1-4 mutant HGMS osgl1-4/oscer1 Acyl-CoA synthetase Pollen coat [94] ago1d mutant TGMS ago1d Argonaute protein PhasiRNA production [77] Table 2.
Types and characteristics of EGMS genes cloned in rice.
-
DGMS line Gene (location) Origin Source Ref. Sanming DGMS *SDGMS/OsRIP1 (Chr.8) Hybrid SE21S × Basmati 370 [98,101,108,109] Pingxiang DGMS Ms-P (Chr.10) Hybrid Pingai 58 × Huaye [97,100] 8987 TMS (Chr.6) Hybrid 3304 × Minghui 63 [96] W450 UK Hybrid Japonica × PGMS [102] OsDMS-2 OsDMS-2A (Chr.2), OsDMS-2B (Chr.8) Inbred 8B1390 [99] zhe9248 M1 UK Artificial mutation Zhe 9248 [103] Orion 1783 UK Artificial mutation Orion [104] Kaybonnet 1789 UK Artificial mutation Kaybonnet [104] OsDMS-1 OsDMS-1A (Chr.1), OsDMS-1B (Chr.2), OsDMS-1C (Chr.3) Tissue culture Zhonghua 11 [105] UK indicates an unknown gene/location. * indicates that the gene has been cloned. Table 3.
The dominant genic male sterile lines and the regulatory genes in rice.
Figures
(6)
Tables
(3)