-
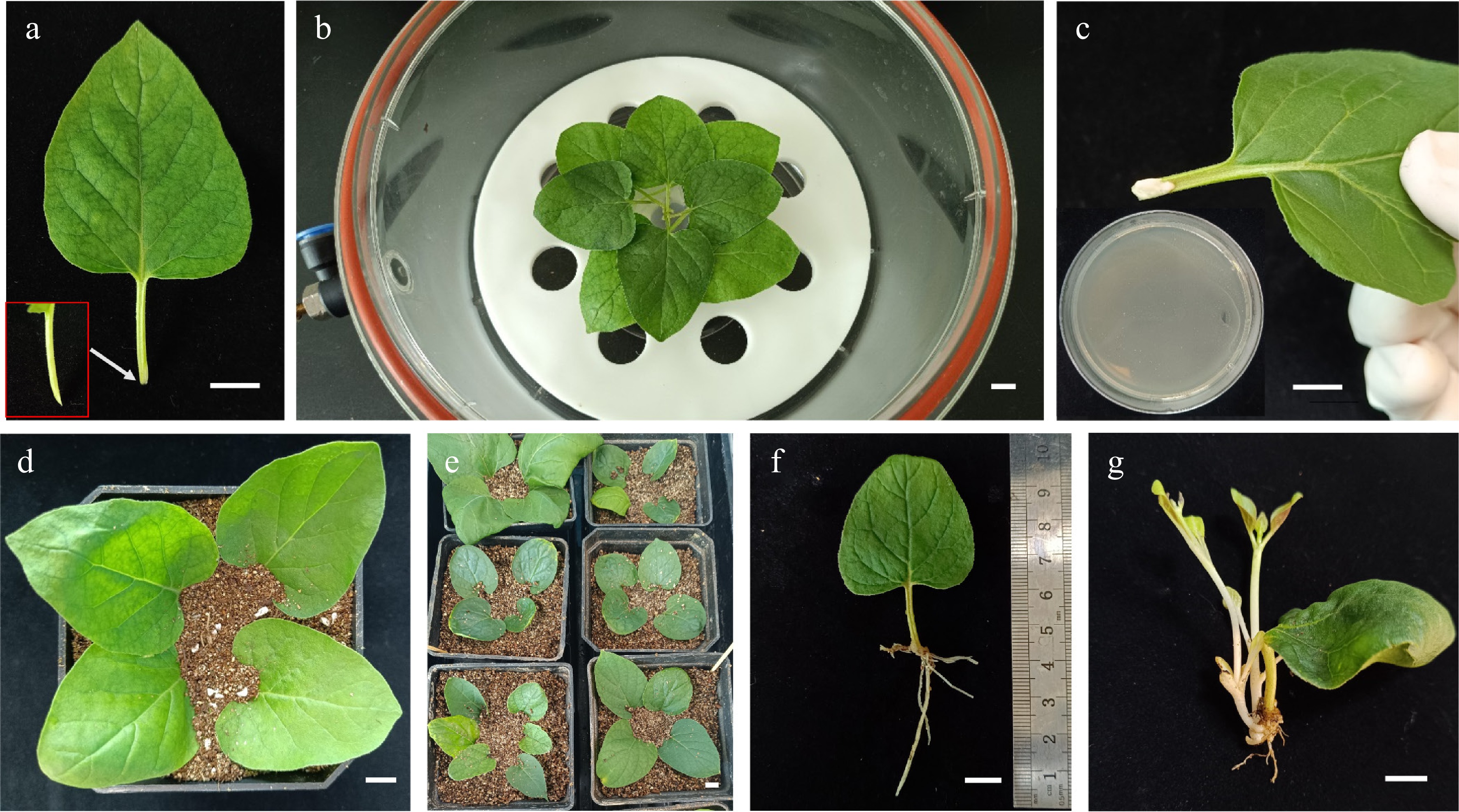
Figure 1.
Genetic transformation of Mirabilis himalaica using the modified CDB protocol. (a) Petiolate leaves were cut and used as explants. The site of infection by A. rhizogene was enlarged. (b) The explants soaked in A. rhizogenes K599 suspension were then subjected to vacuum pressure. (c) Coating the cut sites with A. rhizogenes K599 layers from agar media. (d)−(e) Explants inoculated with A. rhizogenes K599 were cultured in soil. (f) The root of 10 d old explants. (g) Buds emerged after about 30 d. Scale bars: 1 cm.
-
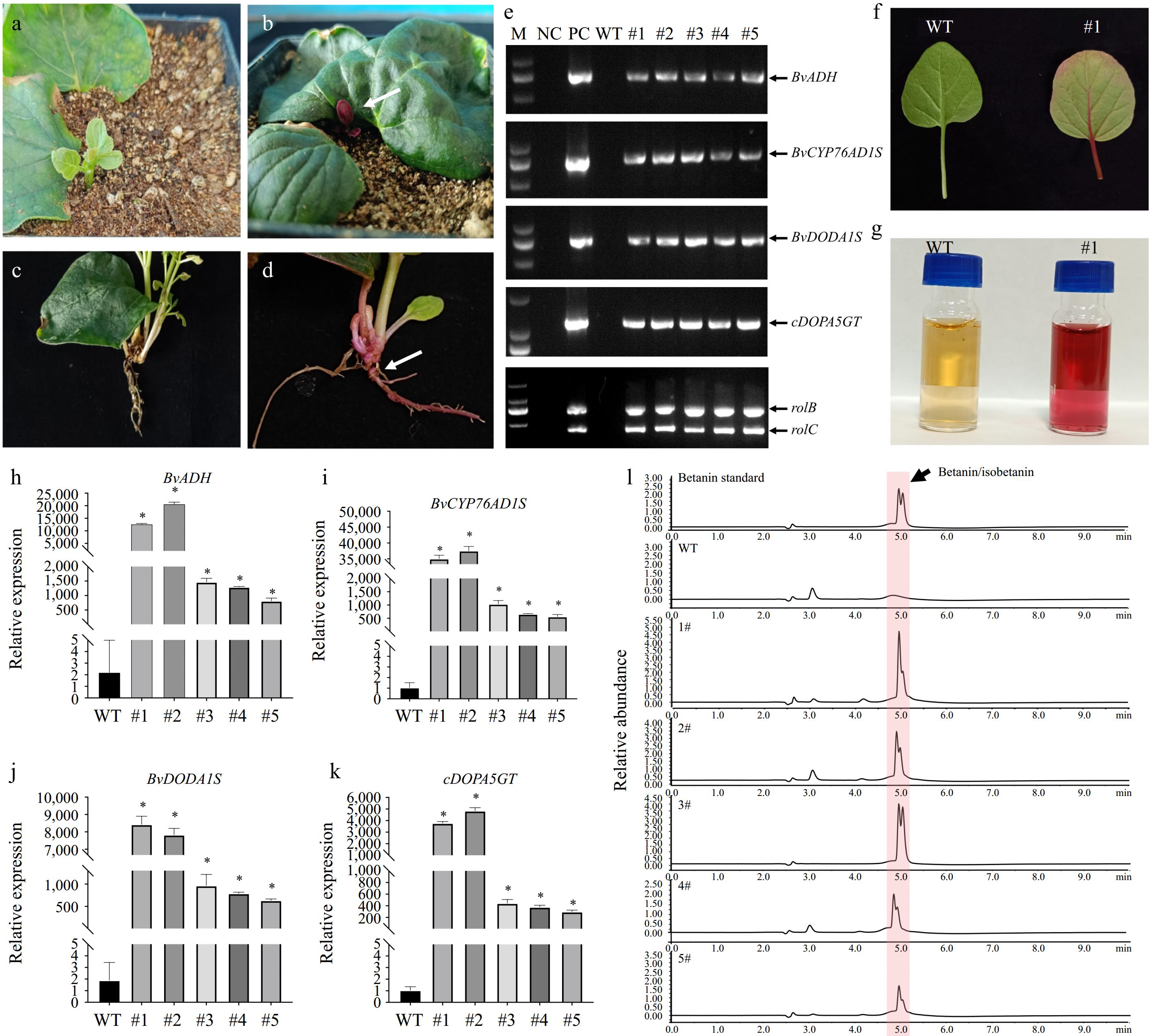
Figure 2.
Genetic transformation of M. himalaica using betacyanin as a reporter. (a) & (b) The above-ground part of M. himalaica, showing the (a) non-transgenic, and (b) transgenic. White arrow indicates red bud with high betacyanin contents. (c) & (d) Roots of M. himalaica. White arrow indicates red roots with high betacyanin contents. (e) Genomic PCR detect four exogenous genes. M: marker; NC: negative control (sterilized water was used as templates); PC: positive control (plasmid pYL1300H-CDGAeG was used as templates); WT: wild type M. himalaica. (f) The betacyanin accumulated in pYL1300H-CDGAeG transgenic M. himalaica leaf. (g) The leaf of pYL1300H-CDGAeG transgenic M. himalaica (right) have higher betacyanin contents compared with wild type M. himalaica (left). (h)−(k) Relative expression levels of betanin biosynthetic genes by qRT-PCR in transgenic M. himalaica, respectively, compared with wild-type (WT) M. himalaica. All data represent the mean ± SD (n = 3; *, p < 0.001; Student's t-test). (l) Analysis of betanin in leaves of M. himalaica by HPLC.
-

Figure 3.
Genetic transformation of M. himalaica using tdTomato as a reporter. (a) & (b) Screening positive plants with a portable fluorescent lamp. Two representative plants growing closely together are displayed here. The plants enclosed in solid rectangular boxes are positive plants and arrow with solid line showed the fluorescent elicited by tdTomato protein. The plants enclosed in dotted rectangular boxes were negative transgenic M. himalaica and the arrow with dotted line indicated negative fluorescent signal. (c) & (d) The fluorescent of positive tdTomato transgenic plants. The roots enclosed in dotted rectangular boxes represented negative transgenic roots and the arrows with solid line showed the fluorescent elicited by tdTomato protein. (e) Genomic PCR detect the tdTomato, rolB and rolC genes in transgenic plants. M: marker; NC: negative control (sterilized water was used as templates); PC: positive control (plasmid Cotton 2.0-tdTomato was used as templates); WT: wild type M. himalaica. (f) Statistics of M. himalaica transformation efficiency using tdTomato as a reporter. Scale bar: 1 cm.
-
Experiment No. of explants No. of buds Ratio of buds/explants I 114 148 1.30 II 135 177 1.31 III 100 132 1.32 Table 1.
Statistics on leaf regeneration and buds emergence of Mirabilis himalaica.
-
Experiment Explants Rooting explants Transgenic roots Transgenic plants Rooting rate Positive rooting rate Positive plant rate I 100 84 75 2 84% 75% 2% II 100 90 80 1 90% 80% 1% III 100 92 79 2 92% 79% 2% Table 2.
Statistics of M. himalaica transformation efficiency using betacyanin as a reporter.
Figures
(3)
Tables
(2)