-
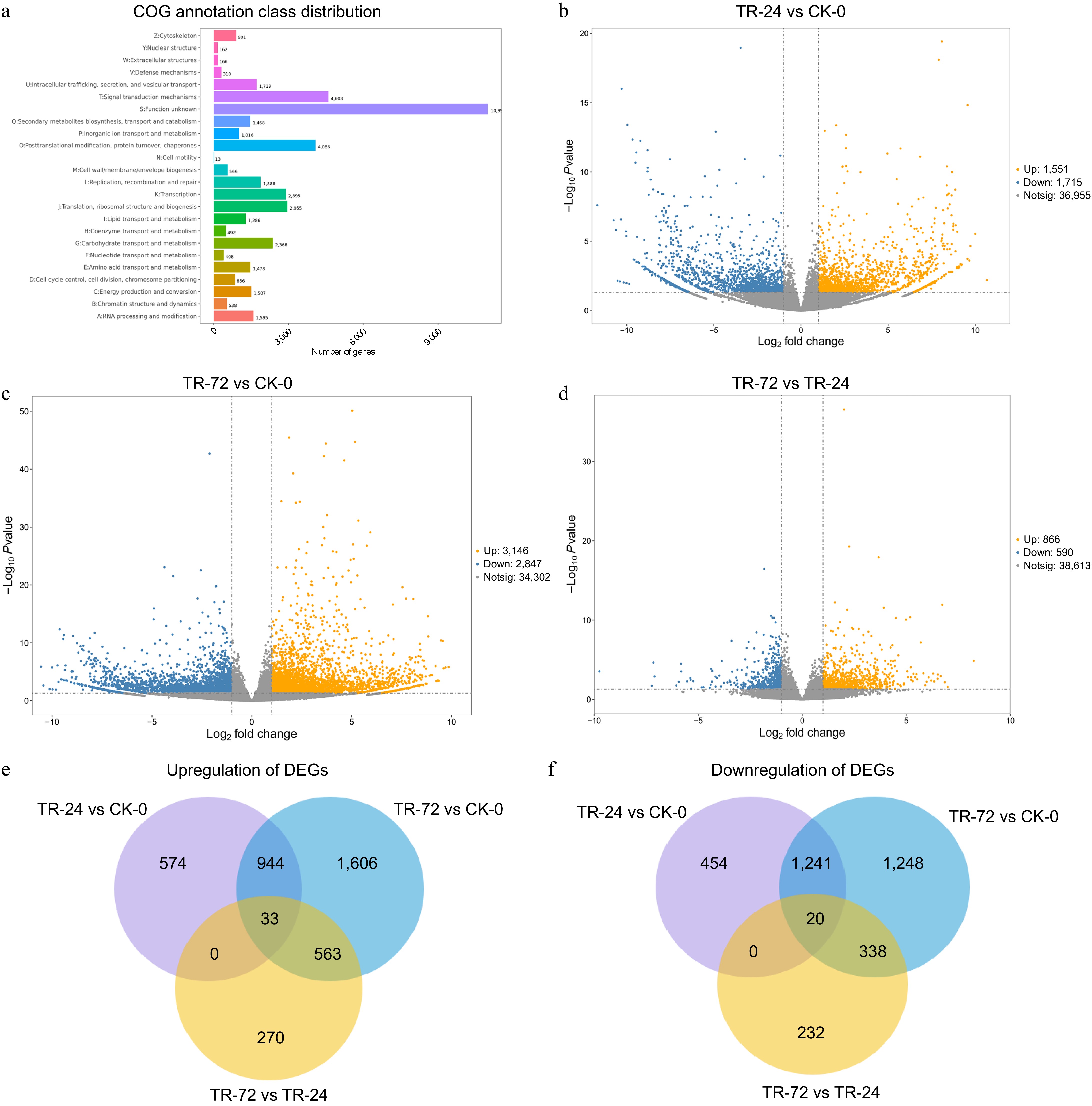
Figure 1.
COG annotation results of DEGs and DEGs in the roots of Chrysanthemum naktongense under lanthanum stress for different durations. (a) COG annotation results of differentially expressed genes. All the predicted genes were consistent with the COG database and were functionally grouped into 24 molecular families. The X-axis represents the number of differentially expressed genes assigned to the corresponding COG class, and the Y-axis represents the COG class to which the differentially expressed genes are annotated. (b)−(d) shows the volcano map of differentially expressed genes in TR-24 vs CK-0, TR-72 vs CK-0, and TR-72 vs TR-24. (e), (f) show Venn diagrams of upregulated and downregulated DEGs in TR-24 vs CK-0, TR-72 vs CK-0, and TR-72 vs TR-24, respectively. The threshold for differentially expressed gene screening was as follows: (|logFC| > 1, p < 0.05).
-
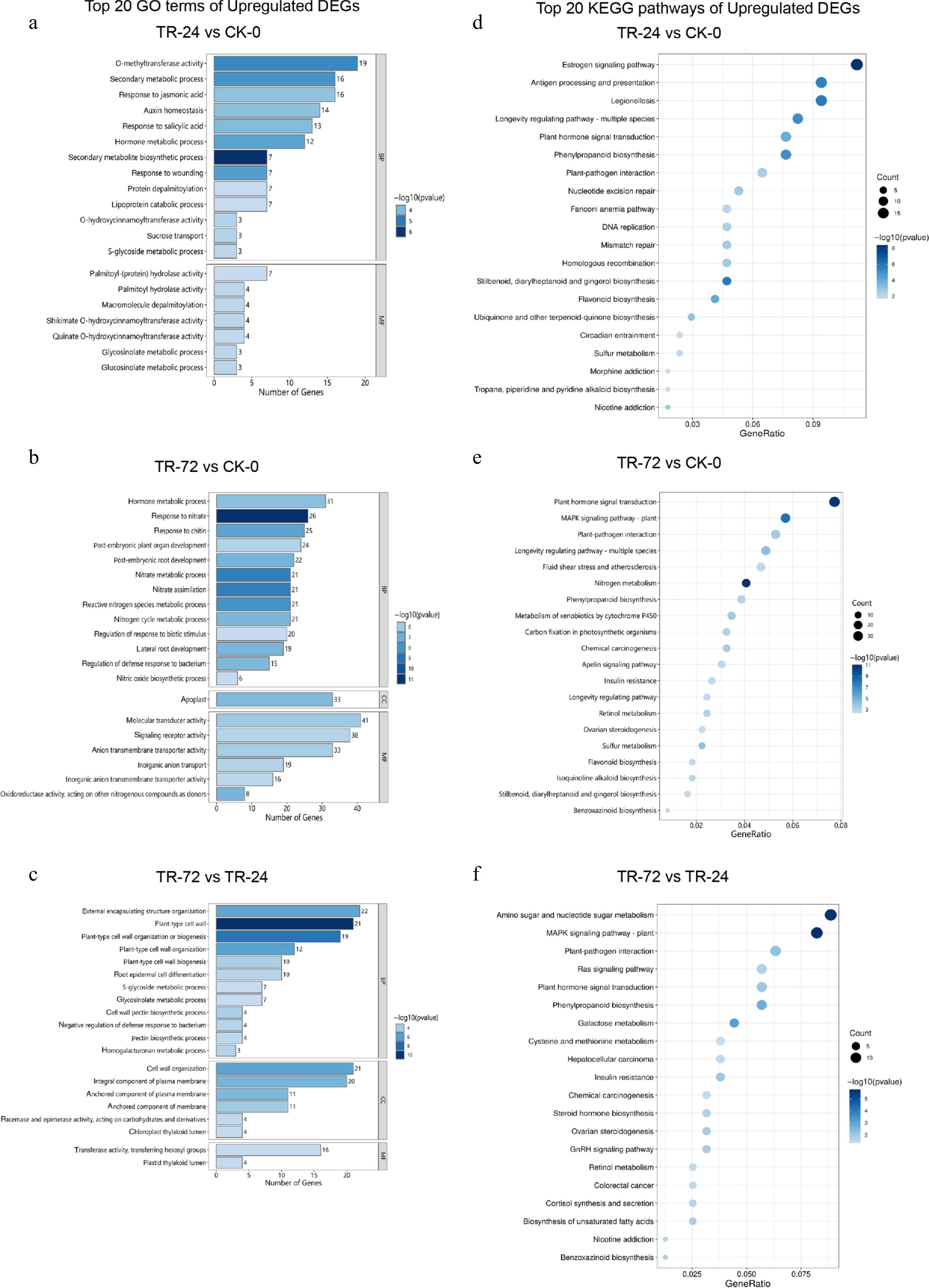
Figure 2.
Column diagram of the top 20 enriched GO terms and bubble diagram of the enriched KEGG pathways of the upregulated DEGs in each treatment group. (a)−(c) Show the top 20 GO enrichment terms in the sequences of TR-24 vs CK-0, TR-72 vs CK-0, and TR-72 vs TR-24. BP refers to biological process, CC refers to cellular component, and MF refers to molecular function. The vertical axis shows the name of the GO enrichment term, and the horizontal axis displays the number of genes enriched in each GO term. The length of the rectangle represents the number of DEGs associated with each enrichment term, and the colour depth represents the significance of each enrichment term. (d)−(f) Shows the bubbles of the TR-24 vs CK-0, TR-72 vs CK-0, and TR-72 vs TR-24 upregulated DEGs in the top 20 enriched KEGG terms. The vertical axis represents the name of the enriched KEGG pathway, and the horizontal axis represents the GeneRatio of each enriched pathway. The size of the points represents the number of DEGs in each enriched pathway, and the depth of the colour of the points represents the significance of each enriched term.
-
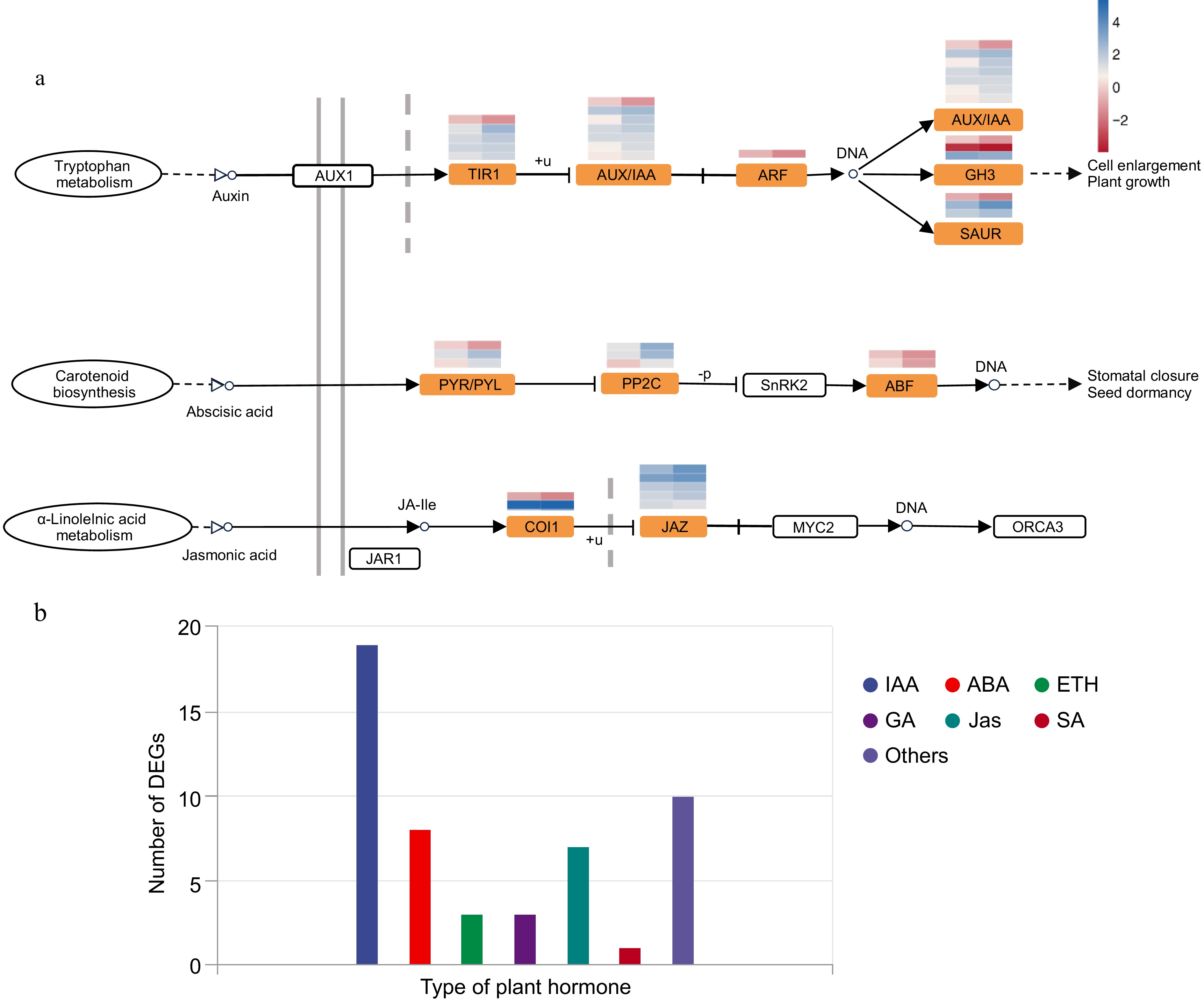
Figure 3.
Plant hormone signal transduction pathway and category of DEGs in this pathway. (a) Expression heatmap of genes associated with the auxin, jasmonic acid, and abscisic acid signal transduction pathways and related enriched DEGs. (b) Number of DEGs annotated to different plant hormones.
-
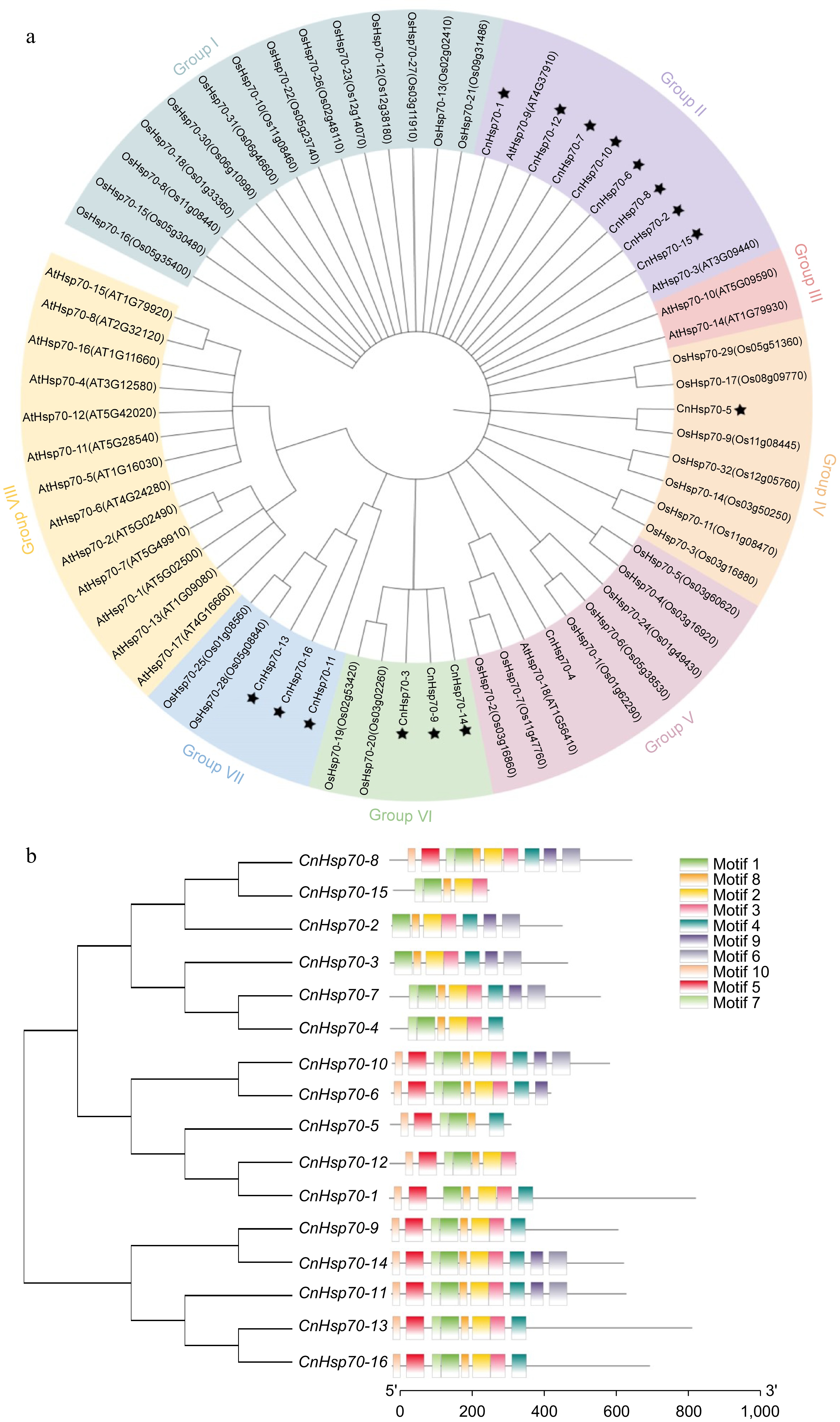
Figure 4.
Phylogenetic tree of Hsp70 family members of Arabidopsis thaliana (At), Oryza sativa (Os), and Chrysanthemum naktongense (Cn) and structure of 16 Hsp70 motifs of Chrysanthemum naktongense. (a) Phylogenetic tree of Hsp70 family members of AtHsp70-1 to 18, OsHsp70-1 to 32, and CnHsp70-1 to 16. Different colors are used to distinguish different groups. The asterisk indicates CnHsp70-1 to 16. (b) Phylogenetic tree of CnHsp70-1 to 16 and their motifs, diverse colors symbolize distinct motifs.
-
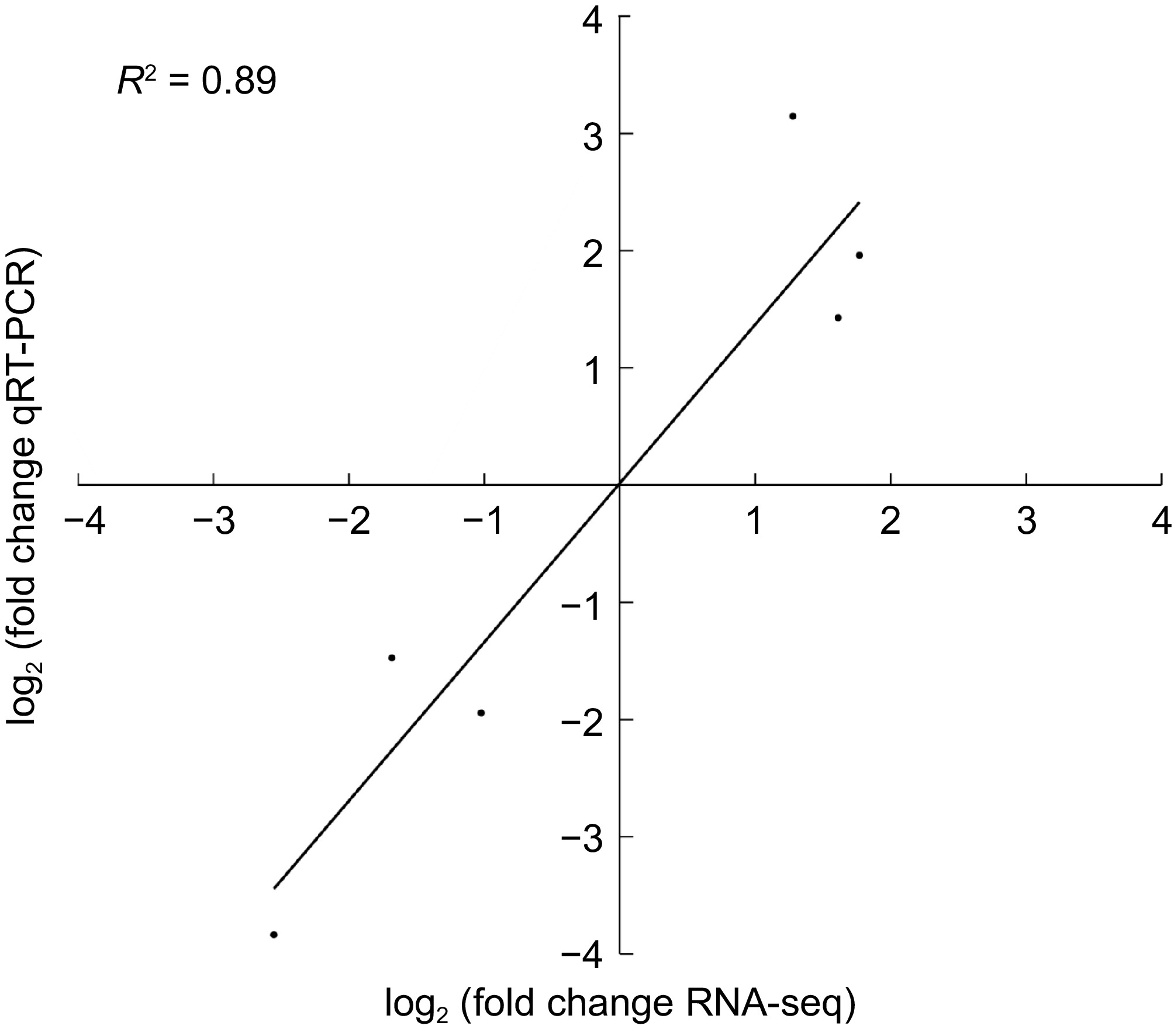
Figure 5.
Six genes were verified via real-time quantitative PCR.
-
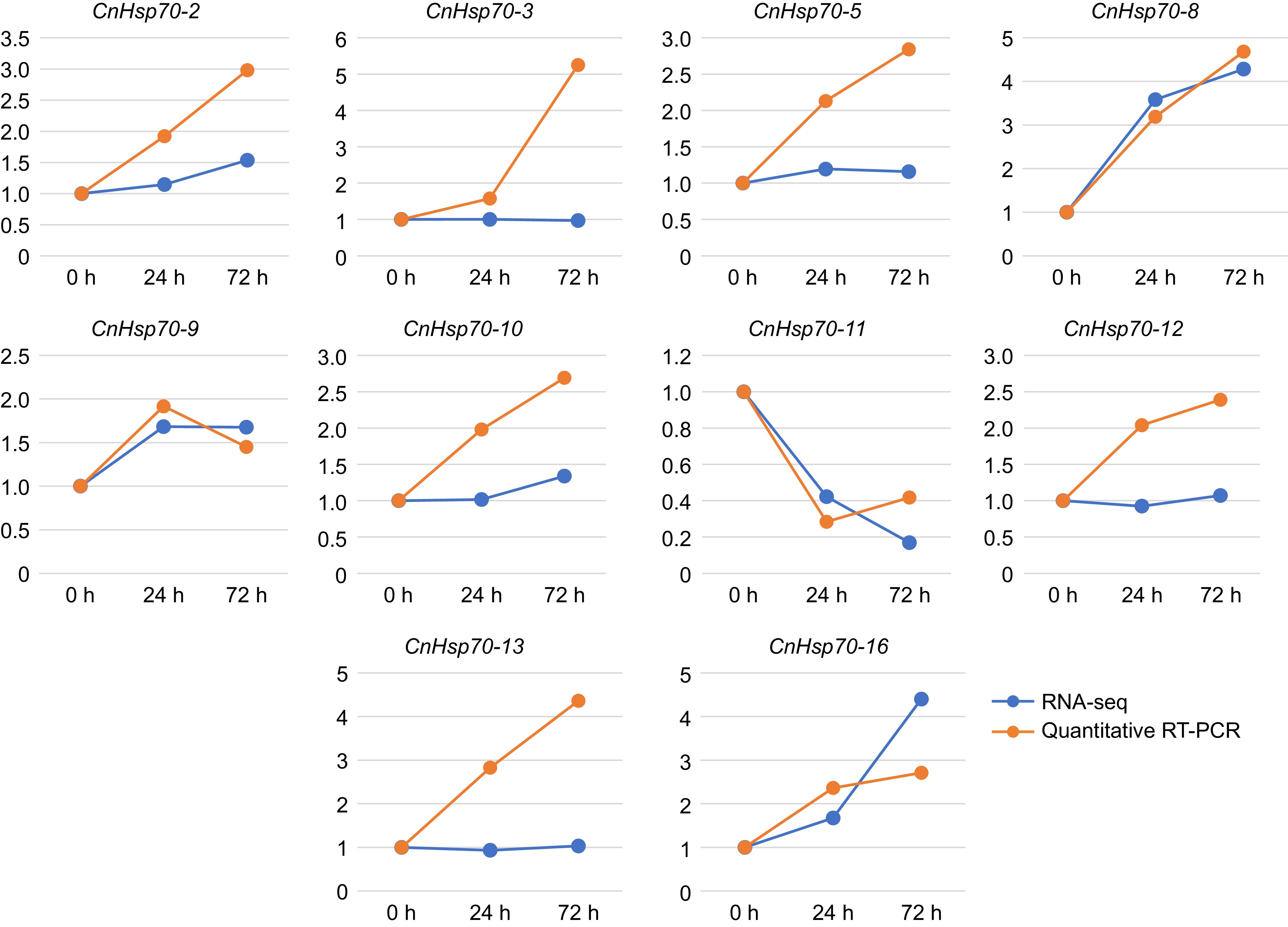
Figure 6.
Expression levels of 10 Hsp70 members of Chrysanthemum naktongense in RNA-seq and qRT-PCR. The blue line represents the results of RNA-seq and the yellow represents qRT-PCR.
-
Sample
nameClean
reads (Mb)Clean
bases (Gb)Data use
rate (%)Q20
(%)Q30
(%)GC
(%)TR-24 (1) 43.90 6.54 98.25 98.73 95.85 43.44 TR-24 (2) 42.33 6.31 98.32 98.60 95.69 44.05 TR-24 (3) 40.70 6.06 98.47 98.66 95.73 43.79 TR-72 (1) 41.68 6.20 98.52 98.81 96.07 43.47 TR-72 (2) 39.38 5.87 98.27 98.78 96.02 43.73 TR-72 (3) 41.94 6.25 98.12 98.70 95.85 43.70 CK-0 (1) 46.22 6.89 98.41 98.67 95.65 43.20 CK-0 (2) 39.77 5.91 97.86 98.69 95.75 44.04 CK-0 (3) 43.33 6.45 98.49 98.79 96.02 43.94 The data use rate (%) was calculated as clean_base/raw_base × 100. Table 1.
Quality of sample sequences.
-
Gene name Gene ID Length (aa) Isoelectric point (pI) Molecular weight (kDa) Instability index Sub-cellular localization Hsp70-1 TRINITY_DN10529_c1_g2_i3 483 6.47 53.17 32.20 Cytoplasmic Hsp70-2 TRINITY_DN10613_c1_g1_i2 493 6.84 55.68 39.82 Cytoplasmic Hsp70-3 TRINITY_DN10917_c0_g1_i7 675 6.02 72.15 36.56 Mitochondrial Hsp70-4 TRINITY_DN152347_c0_g1_i2 614 5.24 66.81 35.86 Chloroplast Hsp70-5 TRINITY_DN152347_c0_g2_i1 352 6.42 39.02 30.19 Chloroplast Hsp70-6 TRINITY_DN16204_c0_g1_i1 335 5.13 36.57 31.48 Extracellular space Hsp70-7 TRINITY_DN1712_c11_g1_i2 265 8.44 29.47 41.39 Chloroplast Hsp70-8 TRINITY_DN1712_c1_g1_i6 585 5.11 64.35 33.28 Cytoplasmic Hsp70-9 TRINITY_DN190619_c0_g1_i1 631 8.71 70.59 50.64 Chloroplast Hsp70-10 TRINITY_DN3151_c0_g1_i18 443 6.63 48.35 37.93 Cytoplasmic Hsp70-11 TRINITY_DN46341_c0_g1_i3 654 5.44 71.27 37.13 Cytoplasmic Hsp70-12 TRINITY_DN5040_c0_g1_i23 864 5.41 96.54 39.62 Plasma membrane Hsp70-13 TRINITY_DN548_c0_g1_i14 832 5.20 92.09 42.99 Cytoplasmic Hsp70-14 TRINITY_DN56386_c0_g1_i1 645 5.13 70.51 37.19 Cytoplasmic Hsp70-15 TRINITY_DN57700_c0_g2_i1 314 8.09 35.75 33.13 Cytoplasmic Hsp70-16 TRINITY_DN756_c0_g1_i18 714 6.31 81.08 42.41 Cytoplasmic Table 2.
The physicochemical characteristics of the 16 CnHsp70 family members.
Figures
(6)
Tables
(2)