-
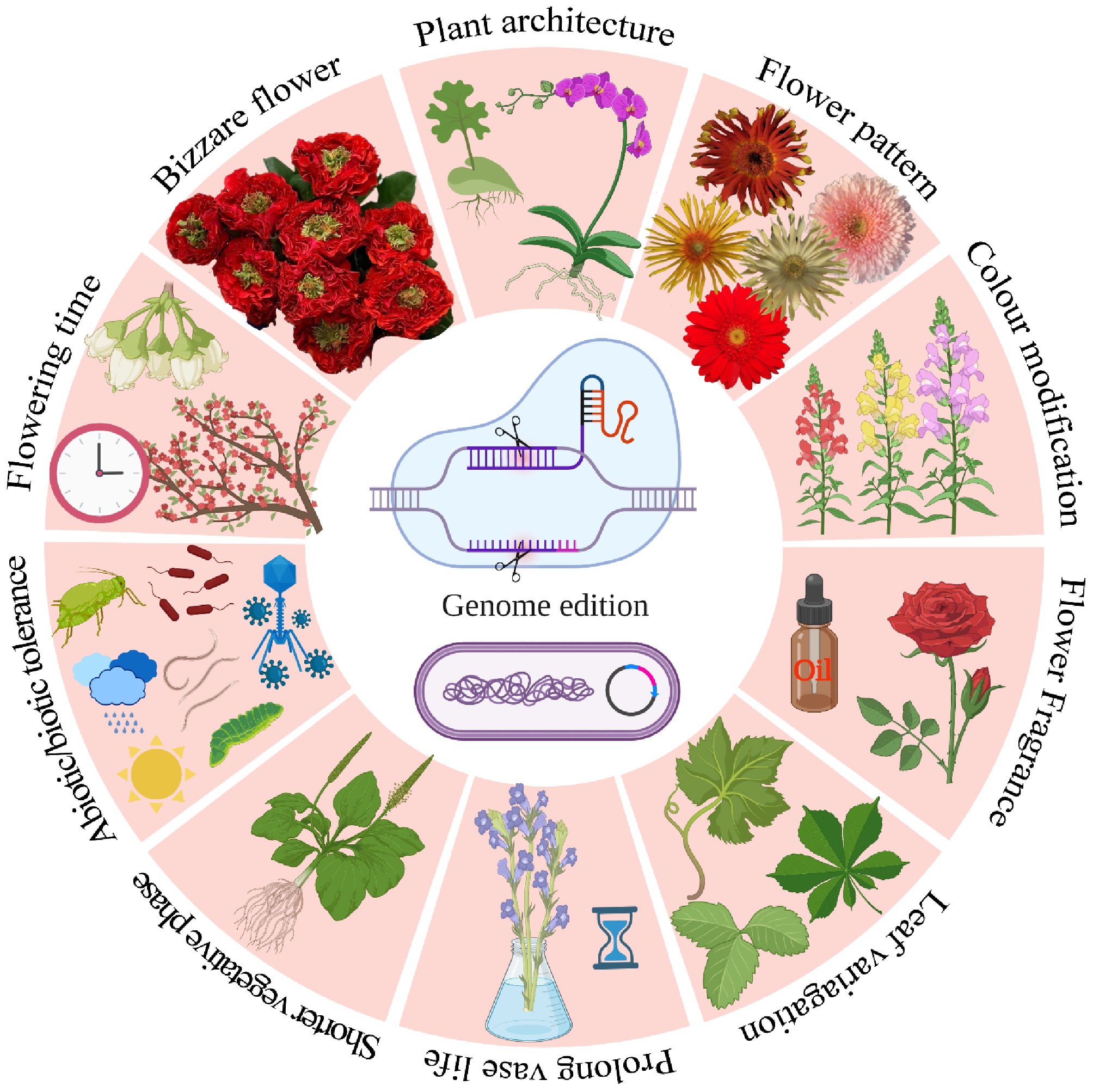
Figure 1.
Applications of genome editing strategies in breeding novel ornamental crops.
-
No. Species Sequencing technology Genome size Chromosome no., and ploidy level Ref. (A) Annuals 1 Amaranthus hypochondriacus Illumina HiSeq 2500 466 Mb 2n = 32 [17] 2 Antirrhinum majus Illumina HiSeq 2000, PacBio RS II 520 Mb 2n = 16 [18] 3 Eschscholzia californica Illumina HiSeq 2500 502 Mb 2n = 12 [19] 4 Gypsophila paniculata MGISEQ-2000 PE 150 5–10 Mb 2n = 2x = 34 [20] 5 Helianthus annuus PacBio RS II 3.6 Gb 2n = 34 [21] 6 Ipomoea nil PacBio, HiSeq 2500 750 Mb 2n = 2x = 30 [22] 7 Mimulus guttatus Illumina HiSeq 2000 531 Mb 2n = 28 [23] 8 Mimosa pudica Illumina HiSeq 2000/4000 896 Mb 2n = 4x = 48 [24] 9 Petunia axillaris HiSeq 2500, PacBio 1.4 Gb 2n = 2x = 14 [25] 10 Petunia inflata HiSeq 2500, PacBio 1.4 Gb 2n = 2x = 14 [25] 11 Salvia splendens PacBio RS II; Illumina HiSeq X Ten 711 Mb 2n = 2x = 44 [26] 12 Tarenaya hassleriana Illumina HiSeq 2000 300 Mb 2n = 20 [27] (B) Aquatic 1 Nelumbo nucifera Illumina HiSeq 2000 879 Mb 2n = 16 [28] 2 Nymphaea colorata PacBio RS II, Hi-C 433 Mb 2n = 28 [29] (C) Cut flowers 1 Chrysanthemum seticuspe Illumina HiSeq 2000 and MiSeq 3.0 Gb 2n = 2x = 18 [30] 2 Chrysanthemum nankingense Oxford Nanopore, HiSeq 2000, PacBio RS II 3.07 Gb 2n = 18 [31] 3 Chrysanthemum lavandulifolium PacBio RS II platform, Oxford Nanopore 2.60 Gb 2n = 18 [32] 4 Dianthus caryophyllus 'Scarlet Queen' contig N50: 14.67 Mb; BUSCO: 97.15% 636 Mb 2n = 2x = 30 [33] 5 Eustoma grandiflorum PacBio HiF 1.71Gb 2n = 6x = 72 [34] 6 Platycodon grandiflorus Illumina HiSeq 2500 platform 680 Mb 2n = 2x = 18 [35] 7 Primulina huaijiensis Illumina HiSeq 2500/HiSeq X Ten platform 478 Mb 2n = 36 [36] 8 Rosa chinensis Illumina HiSeq 2500, PacBio RS-II 532.7 Mb 2n = 2x = 14 [37] 9 Rosa multiflora Illumina MiSeq and HiSeq 2000 platforms 711 Mb 2n = 2x = 14 [38] 10 Rosa rugosa, PacBio HiFi 382.6 Mb 2n = 14 [39] 11 Rosa wichuraiana 'Basye's Thornless' PacBio HiFi, Oxford Nanopore 530.07 Mb 2n = 14 [40] 12 Rosa roxburghii Illumina HiSeq 2500 480.97 Mb 2n = 14 [41] (D) Cacti 1 Carnegiea gigantea Illumina HiSeq 2000, MiSeq 980.3 Mb–1.3 Gb – [42] (E) Grasses 1 Lolium perenne Illumina HiSeq 2000 99 Gb 2n = 14 [43] 2 Trifolium pratense Illumina HiSeq 2000 420 Mb 2n = 14 [44] 3 Trifolium subterranum Illumina HiSeq 2000 552.4 Mb 2n = 16 [45] 4 Zoysia japonica Illumina HiSeq 340 Mb 2n = 40 [46] 5 Zoysia matrella Illumina HiSeq 2000, MiSeq 423 Mb 2n = 40 [46] 6 Zoysia pacifica Illumina HiSeq 2000, MiSeq 302 Mb 2n = 40 [46] 7 Selaginella lepidophylla PacBio RsII platform, Illumina MiSeq platform 122 Mb 2n = 20 [47] (F) Herbaceous perennials 1 Asparagus setaceus HiSeq X Ten, Hi-C 720 Mb 2n = 20 [48] 2 Drosera capensis Illumina HiSeq 2500 264–293 Mb – [49] 3 Dionaea muscipula PacBio RS II 3.19 Gb – [50] 4 Boea hygrometrica Illumina HiSeq 2000, Roche 454 1.69 Gb – [51] 5 Catharanthus roseus Illumina HiSeq 2000 738 Mbp 2n = 16 [52] 6 Primula veris Hiseq 2000, Miseq, Ion Proton, PacBio 479.22 Mb 2n = 2x = 22 [53] 7 Rhodiola crenulata Illumina HiSeq 2000/4000 420.2 Mb 2n = 38 [54] 8 Tanacetum cinerariifolium HiSeq X, HiSeq 4000 7.1 Gb 2n = 18 [55] (G) Orchids 1 Apostasia shenzhenica Illumina HiSeq 2000, PacBio 471 Mb 2n = 68 [56] 2 Dendrobium catenatum HiSeq 2000 1.1 Gb 2n = 2x = 38 [57] 3 Dendrobium officinale Illumina HiSeq 2000, PacBio RS II 1.27 Gb 2n = 2x = 38 [58] 4 Phalaenopsis aphrodite GAIIX, HiSeq 1.20 Gb 2n = 2x = 38 [59] 5 Phalaenopsis equestris Illumina HiSeq 2000 1.16 Gb 2n = 2x = 38 [60] 6 Cymbidium ensifolium PacBio RSII platform 3.62 Gb 2n = 2x = 40 [61] 7 Cymbidium goeringii PacBio Sequel platform 4.10 Gb 2n = 40 [62] (H) Pot plants 1 Begonia fuchsioides Illumina HiSeq 2000/4000 935 Mb 2n = 2x = 22 [24] 2 Kalanchoe fedtschenkoi MiSeq 260 Mb 2n = 34 [63] 3 Primula vulgaris Illumina HiSeq 2500 474 Mb 2n = 22 [64] 4 Rhododendron simsii PacBio RS II, Hi-C 525 Mb 2n = 26 [65] 5 Sedum album PacBio RS II 305 Mb 2n = 48 [66] (I) Shrubs 1 Chimonanthus praecox Illumina HiSeq 2000, PacBio RS II, HiSeq X, Hi-C 778.71 Mb 2n = 22 [67] 2 Chimonanthus salicifolius Illumina HiSeq 2000, PacBio RS II, Hi-C 835.5 Mb 2n = 22 [68] 3 Forsythia suspensa Illumina HiSeq 2500, Oxford Nanopore 701.40 Mb 2n = 28 [69] 4 Gardenia jasminoides Illumina HiSeq 2000, Oxford Nanopore, Hi-C 547.5 Mb 2n = 22 [70] 5 Hibiscus syriacus HiSeq 2000 1.9 Gb 2n = 4x = 80 [71] 6 Jasminum sambac Illumina HiSeq X Ten platform - 537.99 Mb 2n = 2x = 26 [72] 7 Lavandula angustifolia Illumina HiSeq 2000 870 Mb 2n = 50 [73] 8 Osmanthus fragrans HiSeq X ten, Hi-C 733.5 Mb 2n = 46 [74] 9 Paeonia suffruticosa BGISEQ-500, PacBio RS II 13.66–15.76 Gb 2n = 10 [75] 10 Rhododendron delavayi Illumina HiSeq 2000 697.94 Mb 2n = 26 [76] 11 Rhododendron williamsianum Illumina HiSeq 2000, Hic 532.5 Mb 2n = 2x =26 [77] 12 Rhododendron griersonianum Illumina HiSeq X Ten system 677 M 2n = 26 [78] 13 Rhododendron ovatum PacBio Sequel platform 549 Mb 2n = 26 [79] (J) Trees 1 Bombax ceiba Illumina HiSeq 2000, PacBio RS II 809 Mb – [80] 2 Camptotheca acuminata Illumina HiSeq 2000 516 Mb 2n = 44 [81] 3 Casuarina equisetifolia Illumina HiSeq 2000, PacBio RS II 300 Mb – [82] 4 Casuarina glauca Illumina HiSeq 2000/4000 314 Mb 2n = 18 [24] 5 Cerasus serrulata Illumina X ten, Nanopore, Hic-C 256.65 Mb 2n = 16 [83] 6 Cerasus yedoensis Illumina HiSeq 2000, MiSeq, HiSeq X 690.1 Mb 2n = 16 [84] 7 Cercis canadensis Illumina HiSeq 2000/4000 301 Mb 2n = 2x = 14 [24] 8 Fraxinus excelsior Illumina HiSeq 2000, MiSeq, Roche 454 877.24 Mb 2n = 22 [85] 9 Ginkgo biloba Illumina HiSeq 2000/4000 10 Gb 2n = 24 [86] 10 Handroanthus impetiginosus Illumina HiSeq 2000 557 Mb 2n = 40 [87] 11 Prunus mume Illumina GA II 280 Mb 2n = 16 [88] 12 Prunus yedoensis HiSeq X Ten, PacBio RS II 257 Mb 2n = 16 [89] 13 Liriodendron chinense Illumina HiSeq 2000, PacBio RS II, Bionano 1.8 Gb 2n = 38 [90] 14 Jacaranda mimosifolia PacBio Sequel II Platform, Illumina HiSeq 2500 platform 707.32 Mb 2n = 36 [91] Table 1.
A comprehensive and comparative genomics of ornamental plants, insights into genome size, chromosome number, and sequencing technology.
-
Species Delivery method Material Cas9 promotor sgRNA promotor Target gene Final produce Phenotype Ref. Campanula portenschlagiana Agrobacterium In vitro grown petioles CaMV 35S AtU6-26 CpEil1a, CpEil1b Plants Increased ethylene tolerance [141] Chrysanthemum morifolium 'Sei-Marin' Agrobacterium Young leaves PcUbi AtU6 CpYGFP Plant Decreased fluorescence [150] Chrysanthemum morifolium Agrobacterium leaf discs CaMV 35S AtU6-26 CmPDS, CmTGA1 Plant Display an albino phenotype [146] Crocus sativus Agrobacterium Corms CaMV 35S − − Plant − [151] Dendrobium officinale Agrobacterium Protocorms CaMV 35S OsU3 C3HC4H, 4CL, CCR, IRX Plant − [127] Dianthus caryophyllus CRISPR/Cas9 ribonucleoprotein (RNP) complex Leaf segments CaMV 35S AtU6-26 ACS1, ACO1 Plant Reduced ethylene production, delayed flower senescence, improved longevity [140] Euphorbia pulcherrima (Poinsettia) Agrobacterium mediated Internode stem F3′H Flower colour changed from vivid red to vivid reddish-orange [152] Fortunella hindsii Agrobacterium Seedlings CaMV 35S AtU6 FhPDS Plant Albino leaves [153] Ipomoea nil 'AK77' Agrobacterium Embryos PcUbi AtU6g InCCD4 Plant Altered flower color [135] Ipomoea nil 'AK77' Agrobacterium Embryos PcUbi AtU6g InEPH1 Plant Extended flower longevity [138] Ipomoea nil 'Violet' Agrobacterium Embryos PcUbi AtU6g InDFR Plant Altered flower color [132] Ipomoea nil 'Violet' Agrobacterium mediated Immature embryo EPH1 Delay in petal senescence [138] Japanese Gentian (Albireo) Agrobacterium mediated Leaf Gt5GT, Gt3'GT, Gt5/3'AT Pale red violet, dull pink, and pale mauve flowers [154] Japanese Gentian (Albireo) Agrobacterium mediated Leaf GST1 Reduced anthocyanin in petals, white and pale blue flower [136] Lilium longiflorum 'White Heaven' Agrobacterium Seedling scales Ubi OsU3 LlPDS Plant Albino leaves [129] Lilium pumilum Agrobacterium Embryos Ubi OsU3 LpPDS Plant Albino leaves [129] Lotus japonicus Agrobacterium mediated LjLb1, LjLb2, LjLb3 Nodule formation in the presence of rhizobia and establishing low free oxygen concentration in nodule [142] Petunia hybrida 'Madness Midnight' PEG Protoplasts − − PhNR Protoplasts − [143] Petunia hybrida 'Madness Midnight' PEG Protoplasts − − PhF3H Protoplasts Flower color [134] Petunia hybrida 'Mirage Rose' Agrobacterium Young leaves CaMV 35S AtU6 PhACO1 Plant Extended flower longevity [137] Petunia hybrid 'Mitchell Diploid' Agrobacterium mediated Leaf DPL Absence of the Vein associated anthocyanin pattern [155] Petunia hybrida 'Mitchell' Agrobacterium Young leaves CaMV 35S AtU6 PhPDS Plant Albino leaves [144] Petunia × hybrida 'Mitchell Diploid' Agrobacterium Young leaves CaMV 35S 6 (AtU6) PhFT1, PhTFL1a, etc. Plant Early Flowering, Increased Branching [130] Petunia inflata Agrobacterium Young leaves CaMV 35S AtU6 PiSSK1 Plant Altered compatibility of pollens [145] Phalaenopsis amabilis Agrobacterium mediated PDS3 Protocorms Albino leaf [156] Phalaenopsis equestris Agrobacterium mediated MADS44, MADS36 and MADS8 Long juvenile period [128] Populus tomentosa Carr. (clone 741) Agrobacterium mediated Leaf PtoPDS Albino phenotype was observed [157] Festuca arundinacea. Transient Agrobacterium mediated transformation Leaf SGR Chlorophyll degradation [158] Salvia miltiorrhiza (SmCPS1) Tanshinone biosynthesis [133] Torenia fournieri 'Crown Violet' Agrobacterium Young leaves CaMV 35S AtU6 TfF3H Plant Altered flower color [131] Torenia fournieri TfRAD1 Leaf sections Flowers with abnormal shape & pigmentation [159] Table 2.
Comprehensive list of genome-edited ornamentals plants, targets, tools, and phenotypic outcomes.
Figures
(1)
Tables
(2)