-
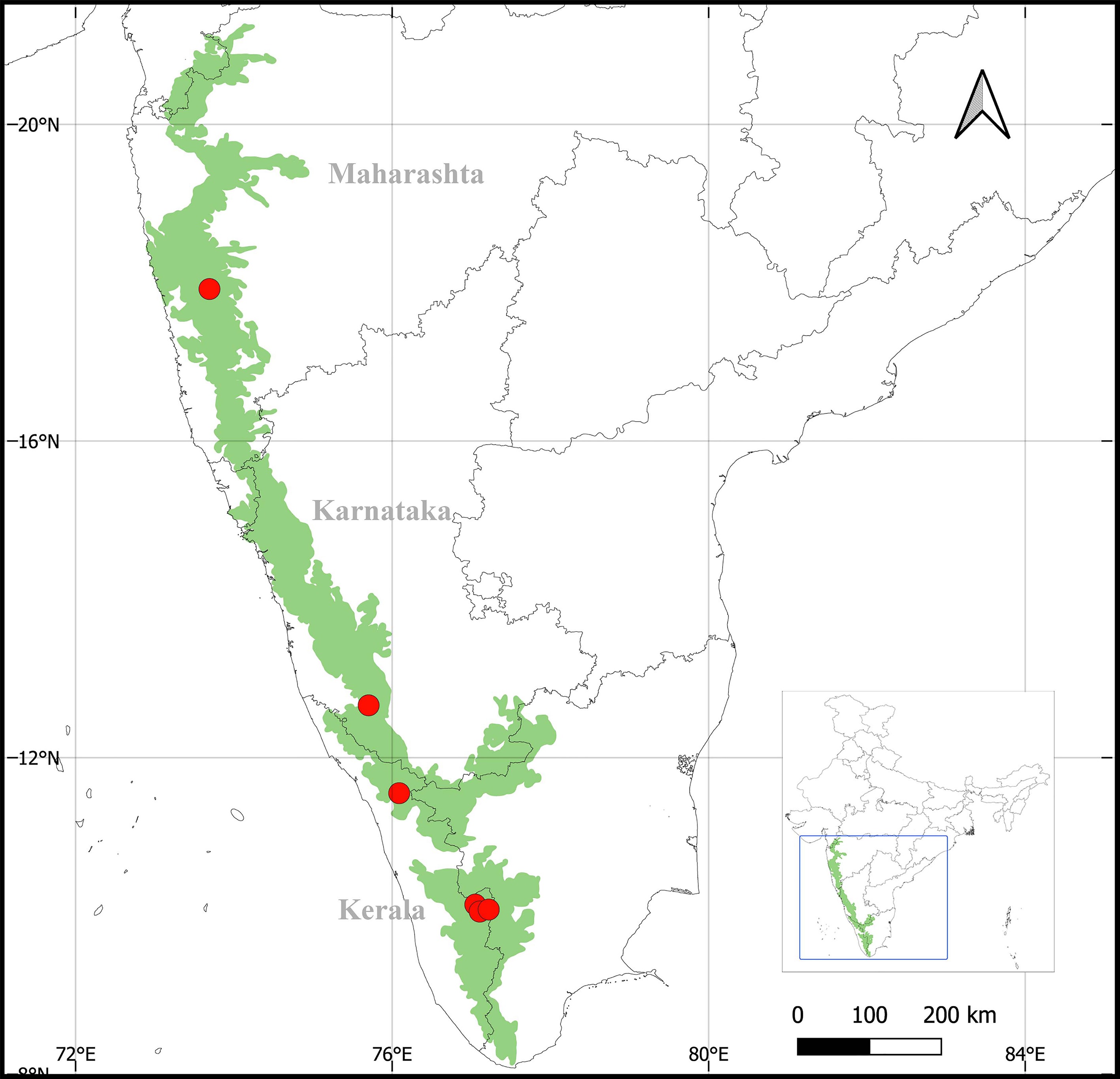
Figure 1.
Species distribution map of Diorygma aeolum in the WGs of India.
-
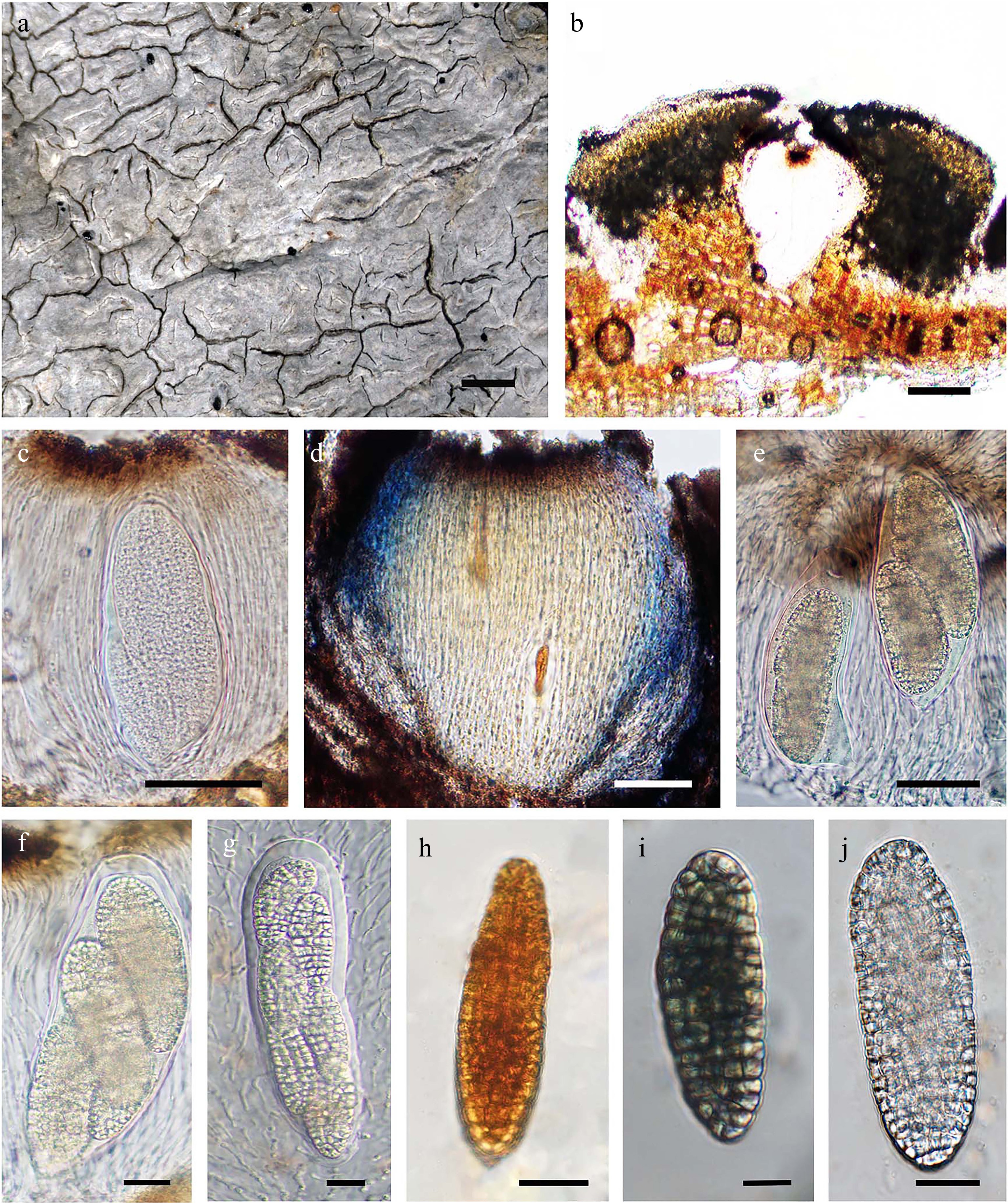
Figure 2.
Diorygma aeolum (AMH22.257). (a) Thallus surface. (b) Lateral section (LS) of lirellae. (c) Hymenium not-inspersed. (d) Hymenium laterally I+ blue in Lugol's iodine. (e)–(g) Asci with a varied number of ascospores. (h) I– ascospore. (i) I+ blue ascospore. (j) Ascospore in water. Scale bars: (a) = 1 mm, (b) = 100 μm, (c) = 10 μm, (d), (e) = 50 μm, (f)–(h) = 20 μm, (i), (j) = 10 μm.
-

Figure 3.
Diorygma albovirescens (holotype). (a), (b) Holotype herbarium material. (c), (d) Thallus. (e)–(j) Ascus showing ascospores. Scale bars: (c) = 1 mm, (d) = 500 μm, (e)–(j) = 20 μm.
-

Figure 4.
Diorygma saxicola (holotype). (a), (b) Holotype herbarium material. (c), (d) Thallus. (e)–(g) Ascus showing ascospores. Scale bars: (c) = 1 mm, (d) = 500 μm, (e), (f) = 100 μm, (g) = 20 μm.
-
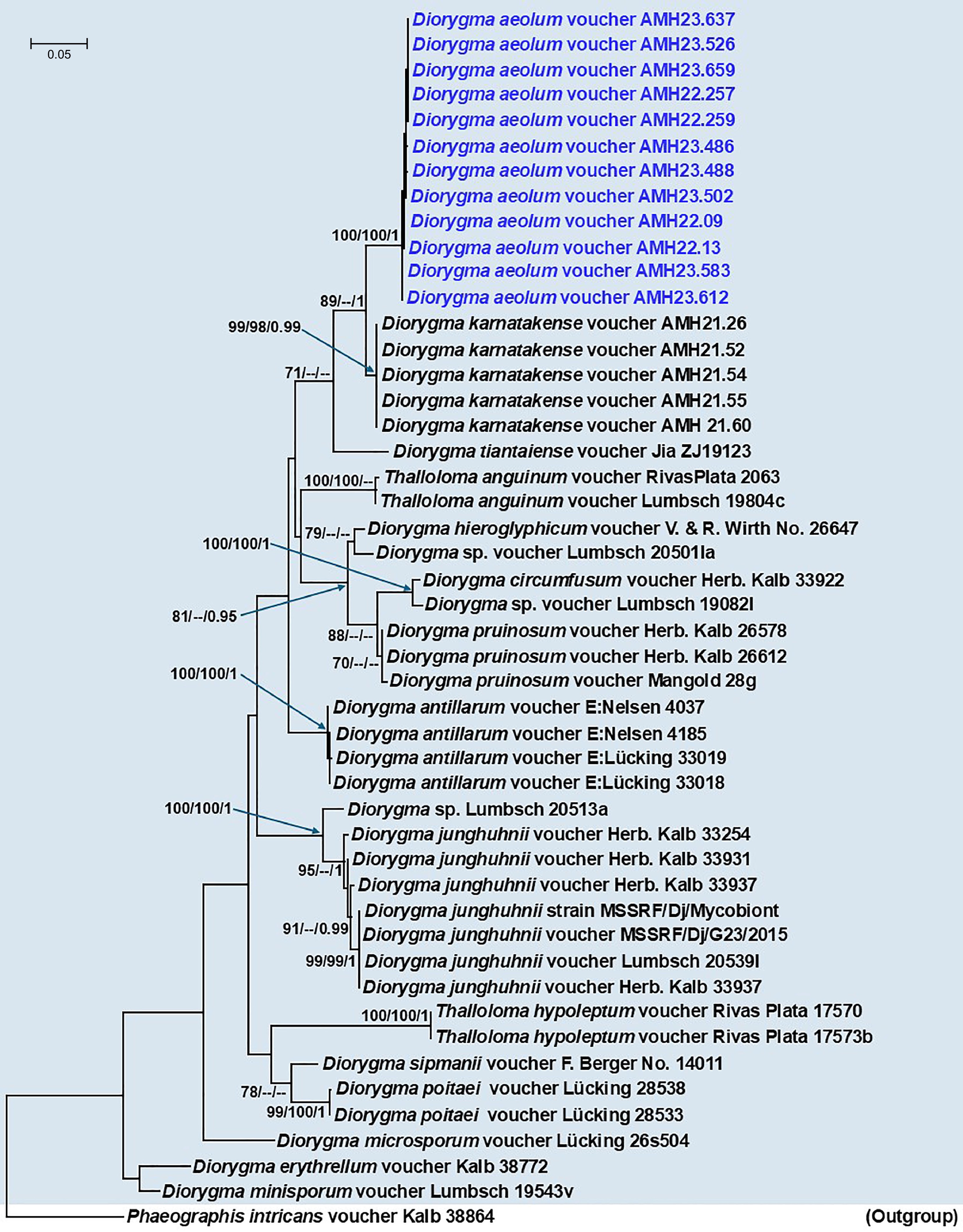
Figure 5.
Phylogenetic tree generated from Maximum Likelihood (ML) analyses based on the combined mtSSU + nuLSU + RPB2 data for the genera Diorygma and Thalloloma species (Graphidaceae). Node support from 1,000 non-parametric bootstraps for RAxML (R-BS), IQ-TREE (UFboot-BS), and posterior probability (PP) from MrBayes are shown at the nodes (R-BS ≥ 70%/Ufboot-BS ≥ 95%/PP ≥ 0.95). Low support values are not denoted. The tree is rooted with Phaeographis intricans voucher Kalb38864. The sequence generated for Diorygma aeolum in this study is indicated in blue.
-
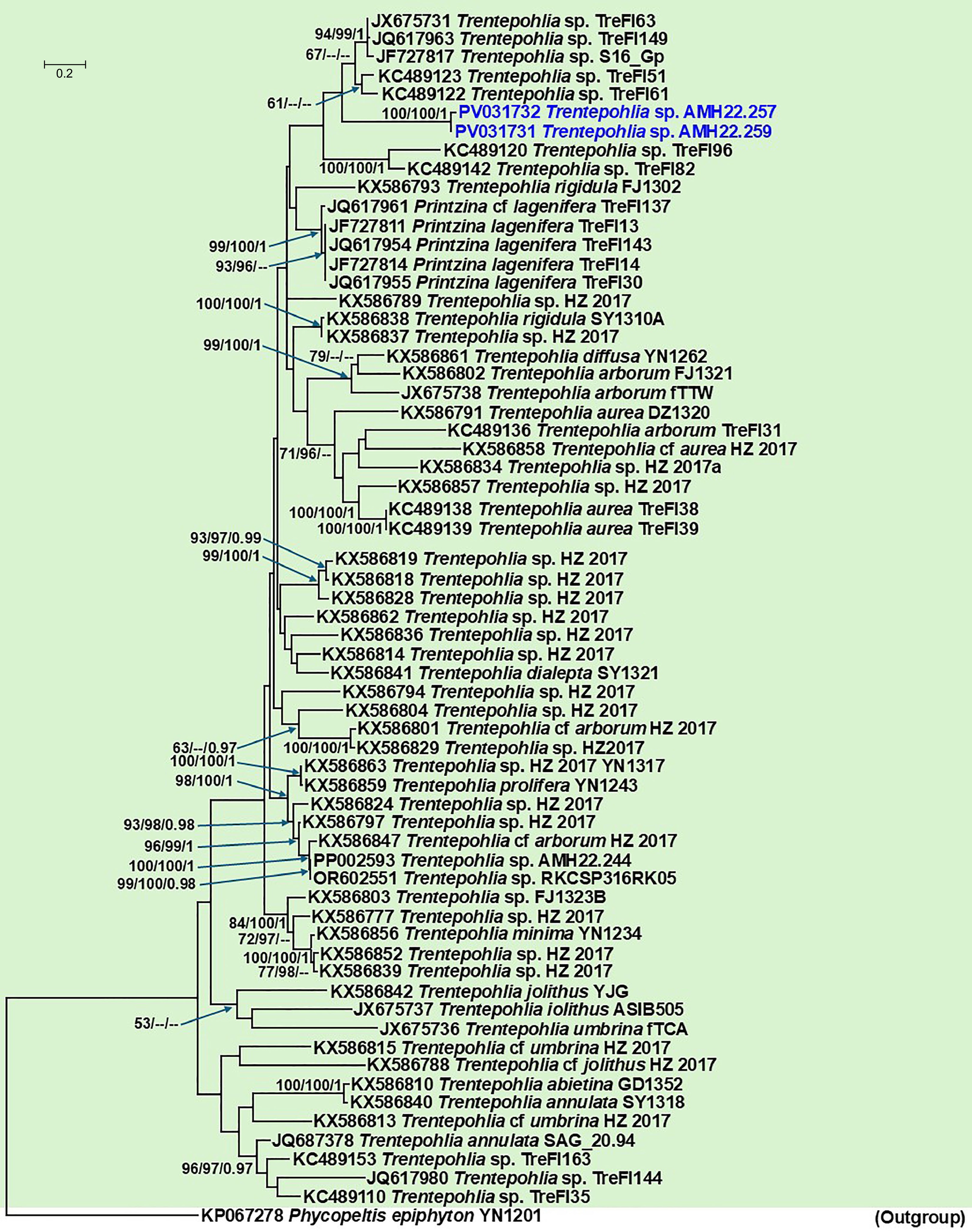
Figure 6.
Phylogenetic tree generated from Maximum Likelihood (ML) analyses based on the combined ITS data for the genus Trentepohlia (Trentepohliaceae), and allied Printzina species. Node support from 1,000 non-parametric bootstraps for RAxML (R-BS), IQ-TREE (UFboot-BS), and posterior probability (PP) from MrBayes are shown at the nodes (R-BS ≥ 70%/Ufboot-BS ≥ 95%/PP ≥ 0.95). Low support values are not denoted. The tree is rooted with Phycopeltis epiphyton voucher YN1201. The sequence generated for Trentepohlia sp. in this study is indicated in blue.
-
Name of taxa Specimen voucher Country mtSSU nuLSU RPB2 Diorygma aeolum AMH22.09 India PV031879 PV034271 − D. aeolum AMH22.13 India PV031880 PV034272 − D. aeolum AMH22.257 India PV031869 PV034274 PV038023 D. aeolum AMH22.259 India PV031870 PV034273 PV038022 D. aeolum AMH23.486 India PV031871 PV034276 − D. aeolum AMH23.488 India PV031872 PV034275 − D. aeolum AMH23.502 India PV031873 − − D. aeolum AMH23.526 India PV031876 − − D. aeolum AMH23.583 India PV031877 − − D. aeolum AMH23.612 India PV031878 − − D. aeolum AMH23.637 India PV031874 − − D. aeolum AMH23.659 India PV031875 − − D. antillarum E:Nelsen 4037 USA JX046452 JX046465 − D. antillarum E:Nelsen 4185 Brazil JX046451 JX046464 − D. antillarum E:Lücking 33019 El Salvador JX046454 JX046467 − D. antillarum E:Lücking 33018 El Salvador JX046453 − − D. circumfusum Herb. Kalb 33922 Australia DQ431963 AY640019 − D. erythrellum Kalb 38772 Thailand JX421022 − − D. hieroglyphicum V. & R. Wirth No. 26647 French Polynesia − AY640015 − D. junghuhnii MSSRF/Dj/G23/2015 India MN944822 − − D. junghuhnii Herb. Kalb 33254 Brazil − AY640016 − D. junghuhnii Herb. Kalb 33931 Australia − AY640017 − D. junghuhnii Herb. Kalb 33937 Australia DQ431962 − − D. junghuhnii MSSRF/Dj/Mycobiont India MN944821 − − D. junghuhnii Lumbsch 20539l Fiji JX421023 JX421474 − D. junghuhnii Herb. Kalb 33937 Australia − AY640018 − D. karnatakense AMH21.26 India OP235521 OP235516 OP245173 D. karnatakense AMH21.52 India OP235522 OP235517 OP245174 D. karnatakense AMH21.54 India OP235523 OP235518 OP245175 D. karnatakense AMH21.55 India OP235524 OP235519 OP245176 D. karnatakense AMH21.60 India OP235525 OP235520 OP245177 D. microsporum Lücking 26504 USA JX421024 − − D. minisporum Lumbsch 19543v Kenya HQ639598 HQ639626 − D. poitaei Lücking 28538 Nicaragua HQ639596 HQ639627 − D. poitaei Lücking 28533 NIcaragua JX421025 JX421475 − D. pruinosum Mangold 28g Australia − JX421476 − D. pruinosum Herb. Kalb 26578 Australia − AY640014 − D. pruinosum Herb. Kalb 26612 Australia DQ431964 − − D. sipmanii F. Berger No. 14011 Costa Rica DQ431961 AY640020 − D. tiantaiense Jia ZJ19123 China − MW750692 − Diorygma sp. Lumbsch 20501la Fiji − JX421478 − Diorygma sp. Lumbsch 20513a Fiji − JX421477 − Diorygma sp. Lumbsch 19082l Australia − JX421479 − Thalloloma anguinum RivasPlata 2063 Philippines JX421337 − − T. anguinum Lumbsch 19804c Fiji JX421336 − − T. hypoleptum Rivas Plata 17570 Philippines JF828970 − − T. hypoleptum Rivas Plata 17573b Philippines HQ639609 − − Phaeographis intricans Kalb 38864 Thailand JX421254 JX421602 JX420924 Newly generated sequences are given in bold. Table 1.
Species name, voucher information, and accession numbers for the mycobiont sequences used in this study.
-
Name of taxa Voucher/isolate/strain information Country ITS Printzina cf. lagenifera TreFl 137 Isolate TreFl 137 United Kingdom JQ617961 Printzina lagenifera TreFl 13 Isolate TreFl 13 Austria JF727811 Printzina lagenifera TreFl 14 Isolate TreFl 14 Italy JF727814 Printzina lagenifera TreFl 143 Isolate TreFl 143 United Kingdom JQ617954 Printzina lagenifera TreFl 30 Isolate TreFl 30 Austria JQ617955 Trentepohlia abietina GD1352 Isolate GD1352 China KX586810 Trentepohlia annulata SAG_20.94 Isolate SAG_20.94 Czehia JQ687378 Trentepohlia annulata SY1318 Isolate SY1318 China KX586840 Trentepohlia arborum fTTW Isolate fTTW Thailand JX675738 Trentepohlia arborum TreFl31 Isolate TreFl31 Brazil KC489136 Trentepohlia arborum FJ1321 Isolate FJ1321 China KX586802 Trentepohlia aurea TreFl38 Isolate TreFl38 Argentina KC489138 Trentepohlia aurea TreFl39 Isolate TreFl39 Argentina KC489139 Trentepohlia aurea DZ1320 Isolate DZ1320 China KX586791 Trentepohlia cf. arborum HZ-2017 Isolate FJ1320 China KX586801 Trentepohlia cf. arborum HZ-2017 Isolate YN1047 China KX586847 Trentepohlia cf. aurea HZ-2017 Isolate YN1240 China KX586858 Trentepohlia cf. jolithus HZ-2017 Isolate DZ1317 China KX586788 Trentepohlia cf. umbrina HZ-2017 Isolate GD1350 China KX586813 Trentepohlia cf. umbrina HZ-2017 Isolate GX1306 China KX586815 Trentepohlia dialepta SY1321 Isolate SY1321 China KX586841 Trentepohlia diffusa YN1262 Isolate YN1262 China KX586861 Trentepohlia jolithus ASIB505 Isolate ASIB505 Austria JX675737 Trentepohlia jolithus YJG Isolate YJG China KX586842 Trentepohlia minima YN1234 Isolate YN1234 China KX586856 Trentepohlia prolifera YN1243 Isolate YN1243 China KX586859 Trentepohlia rigidula FJ1302 Isolate FJ1302 China KX586793 Trentepohlia rigidula SY1310A Isolate SY1310A China KX586838 Trentepohlia sp. AMH22.244 Voucher AMH22.244 India PP002593 Trentepohlia sp. AMH22.257 Voucher AMH22.257 India PV031732 Trentepohlia sp. AMH22.259 Voucher AMH22.259 India PV031731 Trentepohlia sp. HZ-2017 Isolate DZ1318A China KX586789 Trentepohlia sp. HZ-2017 Isolate FJ1305 China KX586794 Trentepohlia sp. HZ-2017 Isolate FJ1313B China KX586797 Trentepohlia sp. HZ-2017 Isolate FJ1323B China KX586803 Trentepohlia sp. HZ-2017 Isolate FJ1324B China KX586804 Trentepohlia sp. HZ-2017 Isolate GX1304 China KX586814 Trentepohlia sp. HZ-2017 Isolate GX1319 China KX586818 Trentepohlia sp. HZ-2017 Isolate GX1326 China KX586819 Trentepohlia sp. HZ-2017 Isolate GX1332 China KX586824 Trentepohlia sp. HZ-2017 Isolate GX1343 China KX586828 Trentepohlia sp. HZ-2017 Isolate GX1345 China KX586829 Trentepohlia sp. HZ-2017 Isolate SY1302 China KX586836 Trentepohlia sp. HZ-2017 Isolate SY1305 China KX586837 Trentepohlia sp. HZ-2017 Isolate SY1314B China KX586839 Trentepohlia sp. HZ-2017 Isolate YN1206 China KX586852 Trentepohlia sp. HZ-2017 Isolate YN1235 China KX586857 Trentepohlia sp. HZ-2017 Isolate YN1316 China KX586862 Trentepohlia sp. HZ-2017 Isolate YN1317 China KX586863 Trentepohlia sp. HZ-2017 Isolate YN1034A China KX586777 Trentepohlia sp. HZ-2017a Isolate HN1008 China KX586834 Trentepohlia sp. KR-2023a Isolate RKCSP316RK05 India OR602551 Trentepohlia sp. S16_Gp Strain S16_Gp Argentina JF727817 Trentepohlia sp. TreFl 144 Isolate TreFl 144 United Kingdom JQ617980 Trentepohlia sp. TreFl 149 Isolate TreFl 149 United Kingdom JQ617963 Trentepohlia sp. TreFl163 Isolate TreFl163 Costa Rica KC489153 Trentepohlia sp. TreFl35 Isolate TreFl35 Argentina KC489110 Trentepohlia sp. TreFl51 Isolate TreFl51 Brazil KC489123 Trentepohlia sp. TreFl61 Isolate TreFl61 Brazil KC489122 Trentepohlia sp. TreFl63 Isolate TreFl63 Brazil JX675731 Trentepohlia sp. TreFl82 Isolate TreFl82 Brazil KC489142 Trentepohlia sp. TreFl96 Isolate TreFl96 Brazil KC489120 Trentepohlia umbrina fTCA Isolate fTCA Austria JX675736 Phycopeltis epiphyton YN1201 (IHB) Voucher YN1201 (IHB) China KP067278 Newly generated sequences are given in bold. Table 2.
Species name, voucher information, and accession numbers for the algal sequences used in this study.
-
Sequence Organism Country Accession no. Identities Gaps mtSSU Diorygma karnatakense voucher AMH21.54 India OP235523 719/736 (98%) 3/736 (0%) Diorygma antillarum isolate MPN528 El Salvador JX046453 686/731 (94%) 19/731 (2%) Diorygma poitaei isolate DNA3210 Nicaragua HQ639596 685/731 (94%) 23/731 (3%) nuLSU Diorygma sp. XZ-2021a voucher Jia ZJ19123 China MW750692 773/847 (91%) 12/847 (1%) Graphidaceae sp. ETRP-2011 voucher Lücking 19955d1 Thailand JF828975 751/861 (87%) 23/861 (2%) Dyplolabia afzelii voucher AMH22.119 India OR602550 750/860 (87%) 28/860 (3%) RPB2 Diorygma karnatakense voucher AMH21.55 India OP245176 749/799 (94%) 2/799 (0%) Diorygma minisporum isolate CHAR48 Kenya KF875520 665/798 (83%) 0/798 (0%) Diorygma aff. minisporum isolate P14636 South Africa ON492053 647/789 (82%) 0/789 (0%) ITS Trentepohlia sp. isolate TreFl81 Brazil KC489141 429/459 (93%) 11/459 (2%) Trentepohlia sp. isolate TreFl56 Argentina KC489140 434/478 (91%) 29/478 (6%) Trentepohlia sp. HZ-2017 isolate FJ1323B China KX586803 374/441 (85%) 24/441 (5%) Table 3.
Details of closest search results of sequenced regions based on MegaBlast.
-
mtSSU + nuLSU +
RPB2Photobiont
ITSNumber of strains (incl. outgroup) 48 65 Length incl. gaps 2,546 886 Distinct alignment patterns 470 675 Undetermined characters (gaps) 48.57% 31.05% Estimated base frequencies A: 0.296 A: 0.248 C: 0.183 C: 0.273 G: 0.255 G: 0.278 T: 0.266 T: 0.202 Substitution rates AC: 1.00000 AC: 1.00000 AG: 3.85637 AG: 1.36556 AT: 1.96997 AT: 1.00000 CG: 1.96997 CG: 1.00000 CT: 10.84645 CT: 2.98126 GT: 1.00000 GT: 1.00000 IQ-TREE analysis Model (BIC) K2P (mtSSU)
TNe+G4 (nuLSU)
TIM+F+I+G4 (RPB2)TN + F + I + G4 Final likelihood value −8,215.620 −14,901.181 Gamma distribution shape parameter (α) 0.155 0.759 RAxML analysis Model (BIC) GTR G + I GTR G + I Final likelihood value −8,246.352430 −14,886.765071 Gamma distribution shape parameter (α) 1.745954 0.784593 Table 4.
Summary of results of the ML analyses performed.
Figures
(6)
Tables
(4)